Relationship of the Genomic Complexity of Hepatitis C Virus With Liver Disease Severity and Response to Interferon in Patients With Chronic HCV Genotype 1b Infection
Hepatology, March 1999, p. 897-903, Vol. 29, No. 3
Francesc-Xavier López-Labrador1,*,
From the 1Liver Unit, Department of Medecine, Institut d’Investigacions Biomèdiques August Pí i Sunyer (IDIBAPS); 2Microbiology Department, Hospital Clínic, Facultat de Medicina, Universitat de Barcelona, Spain.
ABSTRACT
In patients with chronic Hepatitis C, the influence of the genetic heterogeneity of the Hepatitis C virus (HCV) on the progression of liver disease and on the responsiveness to interferon therapy is a matter of controversy. In this study we evaluated the genetic complexity of HCV by single-strand conformation polymorphism (SSCP) analysis of amplicons from the hypervariable region 1 (HVR1) in 168 patients with chronic genotype 1b HCV infection, of whom 122 received a single course of interferon therapy (3 MU, three times weekly for 6 months). No correlation was observed between the degree of genetic complexity of HCV (indicated by the number of bands in the SSCP assay) and patient age, serum alanine aminotransferase activity, or serum HCV-RNA concentration, measured by competitive polymerase chain reaction. HCV genomic complexity was not related to gender nor to the presumed source of infection. The number of SSCP bands detected in serum samples from patients with chronic hepatitis, either mild (8.1 ± 3.9), moderate (8.0 ± 3.3), or severe (9.2 ± 3.3), and in patients with liver cirrhosis, either compensated (8.0 ± 2.9), decompensated (6.3 ± 2.9), or with superimposed hepatocellular carcinoma (9.5 ± 2.9), was similar. The number of SSCP bands detected in patients with sustained response (7.5 ± 3.9), transient response (8.3 ± 2.9), or no response (8.2 ± 3.6) to interferon administration was similar as well. These observations suggest that the genetic complexity of hypervariable region (HVR1) of HCV, as estimated by SSCP analysis, is not related to the severity of liver injury nor to the type of response to interferon therapy. Thus, information offered by SSCP analysis of HVR1 of HCV in chronic HCV genotype 1b infection does not appear to be useful in the clinical management of these patients. (HEPATOLOGY 1999;29:897-903.) (HEPATOLOGY 1999;29:897-903.)
INTRODUCTION
The Hepatitis C virus (HCV) is an RNA virus that replicates with a high rate of mutation,1 which is particularly evident in the hypervariable region 1 (HVR1) of the N-amino terminal region of the second envelope domain of the viral genome.2,3 Under the influence of environmental factors, continuous viral mutation gives raise to a mixed and changing population of mutants, which is known as quasispecies.4 As in infections with other RNA viruses, the quasispecies nature of HCV5 may have important biological implications concerning viral persistence, pathogenicity, and resistance to antiviral agents. However, attempts aimed to define the relationship between clinical aspects of chronic HCV infection and the genetic heterogeneity of the infecting virus have provided conflicting results.
By sequence analysis of HVR1, greater nucleotide sequence diversity between HCV variants was shown in isolates from patients with more advanced liver disease,6 but this finding has not been confirmed by others.7 Studies based on single-strand conformational polymorphism (SSCP) analysis of HVR1 derived amplicons have also provided controversial data. In 1995, Koizumi et al.8 found that the viral populations were more heterogeneous in patients with hepatic cirrhosis or hepatocellular carcinoma than in those with chronic hepatitis, but other studies did not disclose a clear association between the degree of HCV quasispecies complexity and the histological severity of liver disease.7,9,10
At present, the relationship between the degree of genomic complexity of HCV and the type of response to interferon therapy is unclear. Previous studies of a relatively small series of patients indicated that, in general, poor response to interferon was associated with high genomic complexity of HCV, whereas sustained response was associated with low genomic complexity.11-15 However, data on patients infected with genotype 1b are conflicting because the significant correlation between low genetic complexity of HCV and long-term response to interferon therapy found in some studies8,16 was not observed in others.15,17
In the present study we investigated the genomic complexity of HCV by SSCP analysis of the HVR1 in a large series of patients with chronic HCV genotype 1b infection and chronic liver disease of variable severity, ranging from asymptomatic mild chronic hepatitis to decompensated cirrhosis and hepatocellular carcinoma. Because a substantial proportion of the patients received a course of interferon-![]() , the relationship between genomic complexity of HCV and response to interferon was also investigated.
, the relationship between genomic complexity of HCV and response to interferon was also investigated.
PATIENTS AND METHODS
Patients. One hundred sixty-eight patients with chronic liver disease related to chronic infection with HCV genotype 1b were included in the study. All patients were seropositive for anti-HCV antibodies by a third-generation enzyme-linked immunosorbent assay (ELISA) technique (HCV ELISA 3.0, Ortho Diagnostic Systems, Raritan, NJ) and had detectable HCV-RNA in serum by nested PCR of the 5’NCR.18 No patient had previously received antiviral or immunosuppressive therapy, nor did any patient have any other potential cause of chronic liver disease, such as current Hepatitis B virus infection (positive test for Hepatitis B surface antigen), excessive alcohol intake (40 g per day for women and 80 g per day for men), or autoimmune or metabolic disorders affecting the liver. Informed consent was obtained in every case, and the Ethics Committee of our Institution approved the study.
The study population included 131 consecutive outpatients with asymptomatic or minimally symptomatic chronic Hepatitis C who were referred for antiviral therapy between January 1995 and June 1996, and 37 randomly selected patients with advanced liver disease who were hospitalized during the same period of time. Among the former, the histological severity of chronic hepatitis was defined as mild in 41 cases, moderate in 45, and severe in 45, according to recently proposed criteria.19 Among the latter, 17 patients had histological evidence of cirrhosis and were classified as having compensated cirrhosis. Of the 37 patients with advanced liver disease, 17 were admitted for treatment of major complications of hepatic cirrhosis, such as ascites, hepatic encephalopathy, or variceal bleeding, and were classified as decompensated cirrhosis; the remaining 20 had evidence of hepatic cirrhosis with superimposed hepatocellular carcinoma and were admitted for diagnostic or therapeutic purposes. The diagnosis of hepatocellular carcinoma was based on ultrasonographic or computed tomography findings and was confirmed by cytological or histopathological examination of liver specimens in all the cases.
Of the 131 patients with uncomplicated chronic hepatitis, 122 received one full course of interferon therapy, consisting of 3 MU injections of recombinant interferon-![]() ,-2b, three times a week for 6 months, and were followed for at least 6 months on no therapy. The severity of liver injury disclosed by pretreatment liver biopsy in treated patients was considered as mild in 41 cases, as moderate in 41, and as severe in 40, of whom 17 had cirrhosis. The response to treatment was evaluated by biochemical (ALT) and virological (HCV-RNA in serum) parameters. The detection limit of the PCR technique used for qualitative detection of HCV-RNA was 100 copies per mL. Sustained response was defined by the presence of normal ALT and undetectable HCV-RNA at the end of the treatment period and at the end of post-treatment follow-up; transient response was defined by the presence of normal ALT and undetectable HCV-RNA at the end of treatment, followed by either biochemical or virological relapse on post-treatment follow-up; and no response was defined by elevated ALT, and or detectable HCV-RNA at the end of treatment.
,-2b, three times a week for 6 months, and were followed for at least 6 months on no therapy. The severity of liver injury disclosed by pretreatment liver biopsy in treated patients was considered as mild in 41 cases, as moderate in 41, and as severe in 40, of whom 17 had cirrhosis. The response to treatment was evaluated by biochemical (ALT) and virological (HCV-RNA in serum) parameters. The detection limit of the PCR technique used for qualitative detection of HCV-RNA was 100 copies per mL. Sustained response was defined by the presence of normal ALT and undetectable HCV-RNA at the end of the treatment period and at the end of post-treatment follow-up; transient response was defined by the presence of normal ALT and undetectable HCV-RNA at the end of treatment, followed by either biochemical or virological relapse on post-treatment follow-up; and no response was defined by elevated ALT, and or detectable HCV-RNA at the end of treatment.
HCV-RNA Extraction and Complementary DNA Synthesis.
Serum samples were collected immediately before the start of interferon therapy or at the time of liver biopsy in patients with chronic hepatitis or compensated cirrhosis and during hospitalization in patients with decompensated cirrhosis or with hepatocellular carcinoma. Sera was aliquoted 2 hours after venipuncture and stored at ![]() 700 C. HCV-RNA was extracted from 180 µL of serum by the acid-guanidinium isothiocyanate phenol-chloroform method.20 Extracted RNA was resuspended in 50µL of diethylpyrocarbonate-treated water. cDNA synthesis was carried out with 300 units of Moloney Murine Leukemia Virus reverse transcriptase (MMLV, GIBCO-BRL, Gaithersburg, MD) for 1 hour at 37°C, using 5 µL of resuspended RNA, random hexanucleotides (Boehringer Mannheim, Mannheim, Germany), and 20 units of ribonuclease inhibitor (RNAsin, Promega, Madison, WI) in a final volume of 25 µL.
700 C. HCV-RNA was extracted from 180 µL of serum by the acid-guanidinium isothiocyanate phenol-chloroform method.20 Extracted RNA was resuspended in 50µL of diethylpyrocarbonate-treated water. cDNA synthesis was carried out with 300 units of Moloney Murine Leukemia Virus reverse transcriptase (MMLV, GIBCO-BRL, Gaithersburg, MD) for 1 hour at 37°C, using 5 µL of resuspended RNA, random hexanucleotides (Boehringer Mannheim, Mannheim, Germany), and 20 units of ribonuclease inhibitor (RNAsin, Promega, Madison, WI) in a final volume of 25 µL.
HCV Genotyping and HCV-RNA Quantitation. HCV genotype was determined by restriction fragment length polymorphism analysis of the 5’NCR as previously described.21,22
HCV-RNA concentration in serum was measured by an in-house-developed competitive PCR method as previously described.23 This method had a dynamic range between 6 × 103 and 6 × 107 copies per mL and measured values correlated well with measurements obtained by Amplicor HCV Monitor test (Roche Diagnostic Systems, Inc., Branchburg, NJ) in serum specimens infected with HCV genotype 1b.
Amplification of the HVR1 by Nested PCR.
The HVR1 of HCV was amplified by a nested PCR procedure with genotype 1b-specific primers derived from the E2 region. Primary PCR was performed with 5 µL of cDNA in a 25 µL final volume with primers HV1 (sense, nt. 1290-1312, 5′-CGC ATG GCT TGG GAT ATG ATG AT-3′) and HV2 (antisense, nt. 2001-2023, 5′-GTG AAC CCA GTG CTG TTC ATC CA-3′). One microliter of the primary PCR reaction was used as template in a secondary PCR round with primers HV3 (sense, nt.1431-1453, 5′-ATG GTG GGT ACC TGG GCT AAG GT-3′) and HV6 (antisense, nt. 1598-1621, 5′-AGG GAA TTC CTG TTG ATG TGC CA-3′) in a 50 µL final reaction volume. Nucleotide positions are according to Okamoto et al.1 Secondary PCR produced a fragment of 190 base pairs (bp) of the E2 gene including the HVR1. Cycling conditions for both primary and secondary amplifications were as follows: 94°C for 5 minutes (94°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 minute) × 35 cycles; 72°C 5 minutes. Final concentrations of reagents used were: MgCl2 1.5 mmol/L, dNTP 200 µmol/L, primers 0.25 µmol/L, and Taq DNA polymerase (GIBCO-BRL, Gaithersburgh, MD) 0.125 units/25 µL reaction volume. To avoid PCR contamination, Kwok and Higuchi general guidelines were strictly observed.24 Four negative and two positive controls were included in each round of amplification. Amplified products were identified in 1.5% agarose gels after ethidium bromide staining.
SSCP Analysis of the HCV Genomic Complexity in the HVR1.
To avoid underestimation of HVR1 variants present in samples from patients with low viremia levels, approximately the same amount of the 190 bp HVR1 amplified DNA products from each sample was subjected to SSCP analysis. The amount of DNA in amplified HVR1 PCR products was estimated by comparison with DNA standards of known concentration after electrophoresis in agarose gels. About 30 ng of double-stranded HVR1 secondary PCR products were denatured with 10 µL of SSCP loading buffer (95% deionized formamide, 0.01% bromophenol blue, 20 mmol/L ethylenediaminetetraacetic acid) at 95°C for 10 minutes and quickly cooled in an ice-water bath during 10 minutes. Samples were immediately loaded into 12% nondenaturing polyacrylamide minigels (Mini-protean II system, BioRad, Richmond, VA) and electrophoresed during 4 hours at 100 volts at room temperature. Bands were visualized on the gel by silver staining25 and the number of SSCP bands was determined by scanning the gels by using the Bioimage Whole-Band analyzer software, version 2.51 (Bioimage System Co, Ann Arbor, MI).
To assess the reproducibility of the assay, a well-characterized sample with a specific SSCP pattern was used as a control in each experiment. One nondenatured and one denatured control was included in each SSCP gel. Only gels with no bands in the nondenatured control and the appropriate number of bands in the denatured control were considered. In addition, testing under the same conditions was carried out in 50 cDNA samples from different serum samples.
To assess the specificity of the assay, SSCP bands were excised from the gel, reamplified as described above, purified, and directly sequenced by using the Thermosequenase Dye Terminator kit (Amersham, Cleveland, OH) in a 310 DNA sequencer (Applied Biosystems, Westerstad, Germany).
The sensitivity of the assay was determined as follows: reciprocal amounts of two selected samples that had a similar concentration of cDNA, but displayed a different SSCP pattern, were serially mixed to obtain an equimolar amount of total cDNA in each mixture. Then, nested PCR of the HVR1 and SSCP analysis were performed in the mixed cDNA samples. The experiment was repeated twice to check the reproducibility of the technique.
To further evaluate the sensitivity of the SSCP assay to detect minor variants, amplified cDNA from representative samples was cloned into TA cloning plasmid pGEM T (Promega, Madison, WI). Competent XL-1 cells were transformed with recombinant plasmids and 10 to 20 randomly selected clones from each isolate were sequenced as described above.
Statistical Analysis. The median number of SSCP bands observed in samples under study was used to define the viral complexity in the HVR1 as low or high. Qualitative data were analyzed by using the ![]() 2 test or the Fisher’s exact test when necessary. Quantitative values were compared by using the Student’s t test, the Kruskal Wallis test or the analysis of the variance when necessary. P values lower than 0.05 were considered significant. All statistical calculations were performed by using the SPSS for Windows, version 6.0 software package.
2 test or the Fisher’s exact test when necessary. Quantitative values were compared by using the Student’s t test, the Kruskal Wallis test or the analysis of the variance when necessary. P values lower than 0.05 were considered significant. All statistical calculations were performed by using the SPSS for Windows, version 6.0 software package.
RESULTS
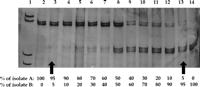
Sensitivity, Specificity, and Reproducibility of the PCR-SSCP Assay. To assess the sensitivity of the PCR-SSCP assay, mixtures containing reciprocal amounts of two samples with a different SSCP pattern were analyzed (Fig. 1). Variants representing 5% to 10% of the whole viral population present in the mixtures were identified as individual bands after silver staining.
|
To View Larger Image |
Fig. 1. Estimation of the sensitivity of the PCR-SSCP assay. Lane 1, molecular weight marker. Reciprocal amounts of cDNA from two isolates (A and B) producing a different SSCP pattern were mixed, amplified by PCR, and submitted to SSCP analysis (lanes 2 to 14). Bands in the upper and lower parts of the gel correspond to HCV variants present in isolate A and in isolate B, respectively. Variants representing at least 5% of the total viral population present in the mixtures produced visible bands in the gel (arrows). |
Analysis of denatured controls showed that the number of bands and the SSCP pattern were identical in repeated experiments, whereas no bands were observed following SSCP of nondenatured controls.
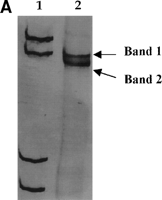
Direct sequencing of reamplified bands excised from the gel was performed to confirm the accuracy of the analysis. Three to four nucleotide changes were observed between two different SSCP bands excised from the same isolate when compared with the consensus sequence (Fig. 2).
|
To View Larger Image Click on Graphic |
Fig. 2. (A) PCR-SSCP analysis of cDNA from isolate A (see Fig. 1). (B) Nucleotide sequences obtained after excision from the gel of the two bands. The sequences obtained from each band differed in three to four nucleotides from the consensus sequence. Dashes represent nucleotide identity with the consensus sequence. Lane 1, molecular weight marker. R = A or G. |
Cloning experiments were carried out by using amplified DNA from samples with low SSCP complexity and from samples with high SSCP complexity (the definitions of SSCP complexity are given below). Sequence analysis of 10 to 20 clones from each isolate showed that the number of individual clones differing from the uncloned sequence in a few nucleotide positions was higher in samples with high than in those with low SSCP complexity (data not shown).
To assess the reproducibility of the method, 50 samples were retested from the cDNA synthesis step. The same numbers of bands and SSCP patterns were observed in all retested samples (data not shown).
HCV Genomic Complexity in the HVR1 and Disease Severity. Nested PCR of the HVR1 generated a single DNA band of the expected size in all the cases. The number of bands detected by SSCP analysis in the whole series ranged from 2 to 20.
The basal features of patients and the results of the SSCP analysis are summarized in table 1. The number of SSCP bands observed in individual cases did not correlate with patient’s age (r = ![]() 0.09, P = .2), serum ALT activity (r = 0.11, P = .14), or HCV-RNA concentration in serum (r = 0.10, P = .3). No differences related to sex were found (8.03 ± 3.3 in men, 8.56 ± 3.7 in women, P = .3). The genomic complexity of HCV did not appear to be related to the presumed route of HCV transmission because the number of SSCP bands was similar in patients with history of blood transfusion (8.7 ± 3.7), in intravenous drug abusers (8.2 ± 3.1), or in those with an unknown source of exposure (7.9 ± 3.2) (P = 0.4).
0.09, P = .2), serum ALT activity (r = 0.11, P = .14), or HCV-RNA concentration in serum (r = 0.10, P = .3). No differences related to sex were found (8.03 ± 3.3 in men, 8.56 ± 3.7 in women, P = .3). The genomic complexity of HCV did not appear to be related to the presumed route of HCV transmission because the number of SSCP bands was similar in patients with history of blood transfusion (8.7 ± 3.7), in intravenous drug abusers (8.2 ± 3.1), or in those with an unknown source of exposure (7.9 ± 3.2) (P = 0.4).
| To View This table | table 1. Basal Features and Complexity of the HVR1, as Determined by SSCP Analysis, in Patients With Genotype 1b HCV Chronic Infection According to the Severity of the Underlying Liver Disease |
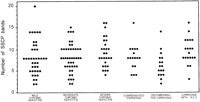
The degree of HVR1 quasispecies complexity in the 114 patients with chronic hepatitis (8.3 ± 3.5) and in the 54 with liver cirrhosis (8.0 ± 3.1) was similar (P = 0.6). Among patients with chronic hepatitis, no significant difference concerning quasispecies complexity between patients with mild, moderate, or severe hepatitis was observed. Similarly, there was no significant difference in the number of observed SSCP bands among patients with compensated or decompensated cirrhosis, or with cirrhosis and hepatocellular carcinoma (table 1 and Fig. 3).
|
To View Larger Image Click on Graphic |
Fig. 3. Relationship between HCV HVR1 complexity, as determined by the number of bands produced by SSCP analysis, and the histological and clinical severity of liver disease in patients with chronic HCV infection. |
To further analyze the possible meaning of quasispecies complexity, patients were separated into two groups with low or high genomic complexity according to the median value of the number of SSCP bands observed in the whole series. Low viral complexity (![]() 8 SSCP bands) was recorded in 103 patients and high viral complexity (> 8 SSCP bands) in 65. The proportion of patients with low or high viral complexity was similar when they were grouped according to clinical or histopathological severity of liver disease (table 1).
8 SSCP bands) was recorded in 103 patients and high viral complexity (> 8 SSCP bands) in 65. The proportion of patients with low or high viral complexity was similar when they were grouped according to clinical or histopathological severity of liver disease (table 1).
HCV Genomic Complexity in the HVR1 and Response to Interferon Treatment.
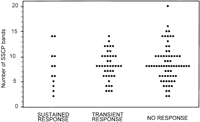
The basal features and the results of SSCP analysis in relation to the observed response to interferon therapy in treated patients are shown in table 2. HCV-RNA serum concentration was significantly lower in sustained responders than in transient responders or in non responders. (P = .04). Differences between these three groups of patients concerning demographic, epidemiological, or biochemical features were not observed. The number of SSCP bands detected in sustained responders, transient responders, and nonresponders was similar (Fig. 4). No significant difference was observed in the proportion of patients with high or low complexity of HVR1 in relation to the type of response to interferon therapy.
| View This table | table 2. Basal Features and Complexity of the HVR1, as Determined by SSCP Analysis, in Patients With Genotype 1b HCV Chronic Infection According to the Response to a Course of Interferon Therapy |
|
To View Larger Image Click on Graphic |
Fig. 4. Relationship between HCV HVR1 complexity, as determined by the number of bands produced by SSCP analysis, and the type of response to one course of interferon- |
DISCUSSION
Host and virus-related factors may influence the natural outcome and the responsiveness to interferon therapy of patients with chronic Hepatitis C. Several studies have shown that HCV genotype and viremia are related to the effectiveness of interferon therapy in these patients,26-28 but the relationship of these factors with the severity of the underlying liver disease is still unclear.
The long-term outcome of HCV infection and its responsiveness to interferon therapy may also be related to the quasispecies nature of HCV, a well-characterized feature of this virus with important biological implications.3,5 In infections caused by many RNA viruses, targets for neutralizing antibodies and for cytotoxic T cells often include epitopes that are located in hypervariable regions with high capacity to change their aminoacid sequence. These changes can provide a better adaptation (fitness) to environmental factors.4 Rapid selection of pre-existent, or emergence of new variants, may confer important biological properties to the virus, including escape from immune clearance, viral persistence, and resistance to antiviral agents.4
In HCV infection, HVR1 escape mutants seem to play a major role in the establishment of persistent infection3 and in the development of resistance to interferon therapy.11-13,29 Sequence analysis of cDNA clones derived from PCR products from individual patients has provided important information on the genetic variability of HCV HVR1.6,7,11,12,17,30,31 However, taking into account the quasispecies nature of HCV and the marked heterogeneity of patients with chronic HCV infection, sequence analysis of a large number of clones would be necessary for accurate assessment of the role of viral heterogeneity in the pathogenesis of the disease and its response to therapy, but this methodology cannot easily be applied to studies involving a large number of patients. This limitation can partially be solved by using alternative indirect methods, such as SSCP analysis of amplified DNA products.32,33 By using this approach, the degree of complexity of the sequences present in a viral population is proportional to the number of bands visible after gel electrophoresis of DNA, and enables a rough estimation of the quasispecies variants present in a given sample. Under the conditions used in the present study, and in agreement with previous observations on the genetic variability of the HCV-NS5A region,34 SSCP analysis allowed detection of quasispecies variants representing as little as 5% to 10% of the whole viral population. Similar results have previously been reported by others.29,35
The possible association between the HVR1 complexity of HCV and the severity of liver disease in chronically infected patients is a conflicting issue. A positive correlation between the degree of HCV genomic complexity and the severity of liver injury has been found in some studies that focused on the hypervariable region6,8 or on the core region of HCV,36 but this finding has not been confirmed by others, either by cloning and sequence analysis7 or by SSCP analysis9,10 of the HVR1 of HCV. However, data from these studies can not be directly compared because of differences in sample size and composition, diversity of HCV genotypes, unequal diagnostic criteria, and no standardization of laboratory methodology. Differences in sensitivity of SSCP analysis may be a crucial point because the number of viral variants in samples from patients with low level of viremia may be underestimated by this technique.10
To overcome these problems, the current study focused on a large and heterogeneous group of patients that included a reasonably high number of subjects with well-defined liver disease at different stages of clinical and histopathological severity. All patients were infected with the same genotype, and approximately the same amount of amplified PCR product was analyzed by using a sensitive and reproducible SSCP technique. In our study, HVR1 genomic complexity was unrelated to patient age and sex, presumed source of infection, ALT serum level, or HCV viremia. Furthermore, no significant differences in the number of SSCP bands were observed between patients with different degrees of liver injury, ranging from asymptomatic mild chronic hepatitis to decompensated cirrhosis and hepatocellular carcinoma. Similar results were obtained when patients infected with isolates with low or high HVR1 complexity were compared. These observations suggest that the width of the mutant spectra of HCV quasispecies, estimated by SSCP analysis, does not appear to be related to degree of liver damage.
Recent data have suggested that HVR1 quasispecies complexity may also be related to the response to interferon therapy. By using SSCP analysis of the HVR1 of HCV, several studies that included a relatively small number of patients infected with a variety of HCV genotypes have shown that the degree of viral complexity was higher in isolates from nonsustained responders than in those from sustained responders to interferon therapy.9,10,13,14 In these studies, however, only a small proportion of the patients infected with HCV genotype 1b developed a sustained response to interferon, and this circumstance makes it difficult to evaluate the role of viral complexity in response to interferon in genotype 1b infected patients. Data from studies that focused on genotype 1b patients are rather conflicting: a good correspondence between low genetic complexity and sustained response to interferon was found in studies from Japan8,16 but not in European patients in whom a relationship between viral complexity and therapeutic response has been observed in genotype 3a, but not in genotype 1b infected patients.15 The reasons for these discrepancies are unclear, but they may be related to differences in patients selection, laboratory methodology or therapeutic schedules.
In the current study, a large number of genotype 1b infected patients who received an identical course of interferon therapy were studied. The number of visualized SSCP bands was similar in long-term responders, in transient responders, and in nonresponders. These data suggest that HVR1 complexity, as evaluated by an accurate SSCP procedure, is not related to the efficacy of interferon therapy. Therefore, SSCP analysis of HVR1 does not appear to be of value as prognostic indicator of response to conventional interferon herapy in patients with chronic hepatitis caused by genotype 1b HCV.
Because the number of HVR1 variants does not appear to be associated with liver disease severity or interferon responsiveness in HCV genotype 1b infection, important questions concerning the pathogenic role of virus-related factors remain unanswered. In the viral quasispecies model, the imposition of either preexisting or emerging mutants with a marked degree of divergence with respect to the global viral population may represent an adaptive advantage for the virus.4 That is, under some circumstances, certain HCV quasispecies variants may be more pathogenic or virulent than others,37 and a single variant bearing drug resistance would survive to the drug more efficiently than multiple variants lacking this specific biological advantage.34 Such particular variants could not be recognized by SSCP and variants present in a very low amount might not be detected. Recently, Polyak et al.,38 by using the heteroduplex tracking assay have suggested that viral diversity in the HVR1, i.e., the genetic divergence between variants, but not viral complexity (the number of quasispecies variants) may be a key factor determining the type of response to interferon. However, it is also possible that mutations in the HCV genome conferring different pathogenicity, virulence, or drug-resistance would be located in genomic regions other than HVR1. If so, variations in the HVR1 would merely reflect the high capacity of this region to accept mutations under different environmental circumstances.
In conclusion, we have found that the genomic complexity in the HVR1 region of HCV genotype 1b is not related to the clinical or histopathological severity of liver disease nor to the type of response to interferon therapy when analyzed by SSCP. Thus, as we recently suggested,34 the long-term outcome of liver disease and the effectiveness of interferon therapy might be related to qualitative, rather than to quantitative features of viral quasispecies variants present in these patients.
Acknowledgment
The authors thank Antoni Llorenç for technical assistance and Llorenç Quintó for help in the statistical analysis.
Abbreviations
Abbreviations: HCV, Hepatitis C virus; HVR1, hypervariable region 1; SSCP, single-strand conformational polymorphism; PCR, polymerase chain reaction; 5’NCR, 5′ noncoding region; ALT, alanine aminotransferase; cDNA, complementary DNA
FootNotes
Received May 17, 1998; accepted October 26, 1998. Present address of F.-X.L.-L.: Division of Gastroenterology, Stanford University School of Medicine, Veterans Administration Medical Center, Palo Alto, CA.
Supported in part by grant SAF 97/0103 from the Comisión Interministerial de Ciencia y Tecnología (CICYT). F.-X. López-Labrador and S. Ampurdanès were supported by Fundació Privada Clínic and M. Giménez-Barcons by Institut d’Investigacions Biomèdiques August Pí i Sunyer (IDIBAPS).
Address reprint requests to: Dr. José M. Sánchez-Tapias, Liver Unit, Hospital Clínic, Villarroel 170, 08036 Barcelona, Spain. E-mail: sanchez@medicina.ub.es; fax: 34-93-4515272.
REFERENCES
Copyright © 1999 by the American Association for the Study of Liver Diseases.