Hepatology, January 1999, p. 217-222, Vol. 29, No. 1
Differences in Hypervariable Region 1 Quasispecies of Hepatitis C Virus in Human Serum, Peripheral Blood Mononuclear Cells, and Liver
Michiari Okuda,
From the First Department of Internal Medicine, Yamaguchi University, School of Medicine, Yamaguchi, Japan.
Hepatitis C virus (HCV) has been reported to potentially replicate in peripheral blood mononuclear cells (PBMCs), but directinformation on the pathogenic implication of HCV infection inPBMCs is still limited. To investigate this issue, we comparedthe complexity of HCV quasispecies in serum, PBMCs, and liversof 13 patients with type C chronic liver disease. Hypervariableregion 1 (HVR 1) was amplified by reverse-transcription polymerasechain reaction (RT-PCR), and the PCR products were subcloned andsequenced. Considerable differences in the complexity of HVR 1 quasispecies were found in the serum, PBMCs, and liver in allpatients, and the predominant sequences from each source weremutually different in 3 (23%) patients. An amino acid sequenceunique to each source existed as well as a sequence common toserum and PBMCs, common to serum and livers, or common to PBMCsand liver. These results suggest infection of PBMCs by HCV, andthat HCV in PBMCs may be differently exposed to host immunityfrom that inliver. (HEPATOLOGY 1999;29:217-222.)
INTRODUCTION
Hepatitis C virus (HCV), a positive-strand RNA virus, is assumed to replicate via the production of a complementary negative-strandtemplate, like that of the closely related flaviviruses.1,2Several lines of evidence suggest that this virus can also infectperipheral blood mononuclear cells (PBMCs) in persistently infectedindividuals on the basis of specific detection of negative-strandHCV RNA in PBMCs by reverse-transcription polymerase chain reaction(RT-PCR) and in situ hybridization3-7; however, this issue isstill controversial.8-10 This has been confirmed in cultured lymphocytes,11-14suggesting that some HCV clones may have an affinity for PBMCs.14However, direct information on the pathogenic implication of HCVinfection of PBMCs islimited.
HCV comprises a heterogeneous mixture of genetically different but closely related variants known as “quasispecies.”15 Themost heterogeneous domains of the HCV genome are present withinthe putative envelope 2 region and are referred to as hypervariableregions (HVR 1 and HVR 2).16,17 Although this quasispeciesnature of HCV is generated by the limited fidelity of RNA replication,host immunity, especially the humoral immune response, is thoughtto be one of the important driving forces in the sequence variationsof the HVR of HCV.18-20 We therefore compared the complexity ofHVR 1 quasispecies of the HCV genome in serum, PBMCs, and liverto assess the pathogenic implication of HCV infection inPBMCs.
PATIENTS AND METHODS
Thirteen Japanese patients with type C chronic liver disease (8 men and 5 women; ages 27-58 years, with a mean age of 47 years)who were positive for antibodies to HCV by a second-generationtest (Ortho Diagnostics, Tokyo, Japan), and had HCV type 1b RNA,were included in this study (table 1). None was homosexual, anintravenous drug abuser, or positive for serological markers ofhepatitis B virus infection or for antinuclear antibodies. Nonereceived any antiviral therapy before the study. Liver biopsywas performed on all patients for diagnostic purposes, and informedconsent in writing was obtained from each patient. Histologicaldiagnoses were made according to recently devised criteria forgrading inflammatory activity (minimal, mild, moderate, and severe)and for staging fibrosis (F1, F2, F3, and F4).21 Four patients(patients 2, 4, 5, and 8) showed minimal histological activityand early fibrosis, 2 (patients 6 and 9) showed mild histologicalactivity and early fibrosis, and 7 (patients 1, 3, 7, 10-13) showedmoderate to severe histological activity and moderate to advancedfibrosis, including cirrhosis. The mean level of HCV RNA in serumwas 106.6±0.4 equivalents per milliliter in all patients except for 1 (patient6) whose HCV-RNA level was below the detection limit.
| View This table | table 1. Clinical Characteristics of Patients |
Serum and PBMC samples were obtained on the same day as liver biopsy. Biopsy specimens were divided into two portions: onewas for light microscopy, and the other was used for detectionof HCV RNA. This second portion was immediately put in acid guanidiniumsolution and frozen in liquid nitrogen. The PBMCs were separatedfrom 10 mL of heparinized blood by density gradient centrifugationwith Leuco PREP (Becton Dickinson, Lincoln Park, NJ), washed threetimes with RPMI 1640 culture medium, and stored at ![]() 80°C untiluse, as were serum and liverspecimens.
80°C untiluse, as were serum and liverspecimens.
Genotyping of HCV RNA
HCV genotypes were determined by a modification of the method of Okamoto et al.,22 as we described previously.23
Measurement of HCV RNA in Serum
The HCV-RNA level in all serum samples was measured by signal amplification with a branched DNA probe assay (Quantiplex, Chiron,Emeryville, CA). The detection limit of the assay is 350,000 (or105.54) HCV genome equivalents permilliliter.
Cross-Mixing Test and Treatment of PBMCs With Trypsin-EDTA and RNase A
To eliminate false positives in PBMC samples caused by contamination by serum HCV RNA, the following experiments were performed:PBMCs obtained from 5 healthy controls were incubated with 3 mLof serum including a high titer of HCV RNA (2.5 × 106 genome equivalents/mL) at room temperature for 30 minutes. Aftercentrifugation, PBMCs were treated with trypsin (final concentration:0.05%) and ethylenediaminetetraacetic acid (EDTA) (final concentration:0.02%), and incubated at 37°C for 20 minutes. Subsequently, 50 µL of RNase A (10 mg/mL; Boehringer Mannheim, Tokyo, Japan) wasadded, and the cell suspension was incubated at 37°C for 15 minutes,as described by Willems et al.4 Thus, PBMCs from healthy controlsthat had been preincubated with HCV-RNA-positive serum as wellas PBMCs from 9 patients (patients 1 to 9) were subjected to RT-PCRfor amplification of the 5′-noncoding region24 with or withouttreatment by trypsin-EDTA and RNaseA.
Detection of Negative-Strand HCV RNA
We used two different experimental approaches, because the methodology for detecting negative-strand HCV RNA is still verycontroversial. One was that of Willems et al.4 (method I),and the other was that of Lanford et al.10 (method II). PBMCsfrom 4 patients (patients 4, 5, 7, and 9 ) were available forboth methods, and were available only for method I from 5 patients(patients 1-3, 6, and 8). Negative strands were detected by synthesisof cDNA with the outer sense primer deduced from the 5′-noncodingregion (5′-CTGTGAGGAACTACTGTCTT-3′).25
Method I. RNA extracted from serum and PBMCs was heated at 70°C for 5 minutes and subsequently kept on ice. cDNA was synthesized usingthe same parameters and conditions as for detection of the positivestrand,24 followed by heating in boiling water for 60 minutesfor inactivation of reverse transcriptase. Subsequently, all sampleswere treated with RNase A at a concentration of 50 µg/mL at 37°Cfor 30 minutes to digest total RNA to prevent possible reversetranscription and subsequent amplification of positive-strandHCV RNA by Taqpolymerase.
Method II. This was performed using the method described by Lanford et al.,10 except for adding second-round PCR. In brief, false primingof the incorrect strand was avoided by conducting cDNA synthesisat 70°C with thermostable rTth DNA polymerase (Perkin-Elmer AppliedBiosystem, Chiba, Japan). Strand-specific amplification was accomplishedby chelating Mn2+ and adding Mg2+, which results in DNA polymerase activity without significantreverse-transcriptase activity. After incubation with RNase Aat 37°C for 15 minutes, second-round PCR was performed using thesame parameters and conditions as for detection of the positivestrand.24 Sera, PBMCs, and liver biopsy specimens from 3 otherpatients, apart from patients 1 to 13, were examined for negative-strandHCV RNA, using method II, as a preliminaryexperiment.
Amplification of HVR 1 by RT-PCR
One hundred microliters of serum, PBMCs (106-107 cells suspended in RPMI 1640), or homogenized liver tissue was mixed with 900 µL of RNAzolTMB (Biotex, Houston, TX). After treatment of PBMCs by trypsin-EDTAand RNase A as described above, RNA was extracted, reverse-transcribed,and DNA fragments containing the HVR 1 were amplified using theprimers for the first-round PCR (sense primer, 5′-GGCCTTGCCTACTATTCCAT-3′;antisense primer, 5′-TGCCAGCAATAAGGCCTCTG-3′), and those for thesecond-round PCR (sense primer, 5′-GGCCTTGCCTACTATTCCAT-3′; antisenseprimer, GTCCTGTTGATGTGCCA-3′), as described previously.26 Theprocedures developed by Kwok and Higuchi27 were employed toavoid false-positiveresults.
Cloning and Sequencing
The PCR products were subcloned into pGEM-T vector (Promega, Madison, WI) by TA cloning, using a DNA Ligation Kit (TakaraBiomedicals, Ohtsu, Japan), as described previously.28 Insertionsof DNA fragments were confirmed by PCR, using universal primers(sense primer, TGTAAAACGACGGCCAGT; antisense primer, CAGGAAACAGCTATGACC).After purification of these PCR products with MicroSpin columns(Pharmacia Biotech, Uppsala, Sweden), 8 to 14 subclones per specimen(serum, PBMCs, or liver tissue) (398 clones total) were sequencedin both directions with an ABI 373S sequencer, using a Dye terminatorcycle sequencing ready reaction kit with AmpliTaq DNA polymerase,FS (Perkin Elmer, Foster,CA).
All data were expressed as the mean ± SD. ANOVA was used to compare the complexity of HVR 1 quasispecies in serum, PBMCs,and liver, defined as the number of distinct amino acid sequencesfrom eachsource.
Informed consent in writing was obtained from each patient, and the study protocol conformed to the ethical guidelines ofthe 1975 Declaration of Helsinki and was approved by the institutionalethicscommittee.
RESULTS
PBMCs from healthy controls were all negative for HCV RNA after incubation with HCV-RNA-positive serum and treatment withtrypsin-EDTA and RNase A, but those from 1 healthy control werepositive for HCV RNA without treatment. In contrast, PBMCs from9 patients were all positive for HCV RNA irrespective of treatmentwith trypsin-EDTA and RNase A (Fig. 1).
| Fig. 1. Detection of HCV RNA in PBMCs from healthy controls and patients. The expected size of PCR products is 145 bp. N, negative control; P, positive control. Numbers between photographs indicate those of healthy controls or patients (patients 1 to 9). |
Using method I, negative-strand HCV RNA was detected in PBMCs in 7 patients (patients 1-4, 7-9), but was not detected in serumin any patient. In a preliminary experiment with method II, negative-strandHCV RNA was detected from liver in 2 of 3 patients, but not fromserum or PBMCs in any patient. Similarly, negative-strand HCVRNA was not detected from serum or PBMCs in any patients examined(patients 4, 5, 7, and 9), using method II (table 2).
| View This table | table 2. Detection of Negative-Strand HCV RNA in Serum PBMCs, and Liver |
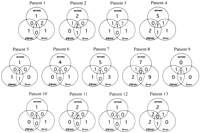
Differences in the complexity of HVR 1 quasispecies were found between serum, PBMCs, and liver in all patients, and the predominantclones from each source were mutually different in 3 patients(patients 8, 9, and 13) (Fig. 2). The total number of distinctamino acid sequences obtained from serum, PBMCs, and liver rangedfrom 3 (patient 10, 10a-10c) to 19 (patient 8, 8a-8s). The percentagedifference in amino acid sequences between the predominant cloneand another minor clone ranged from 3.7% to 59.3% (Fig. 2). Thecompositions of HVR 1 quasispecies in serum, PBMCs, and liverare illustrated in Fig. 3. Amino acid sequences common to serumand PBMCs were found in 7 patients (patients 2, 3, 7-9, 11, and13); those common to serum and liver were found in 5 patients(patients 1, 3, 4, 9, and 13); those common to PBMCs and liverexisted in 5 patients (patients 2, 4, 5, 9, and 13). Amino acidsequences unique to serum existed in all patients except for patient9; those unique to PBMCs or liver existed in 8 patients (patients1, 3-5, 7, 8, 12, and 13), and in 10 patients (patients 1-4, 6-8, 10-12), respectively. The complexity of HVR 1 quasispecies, definedas the number of distinct amino acid sequences, was 4.7 ± 2.1 in serum (P = .0084 vs. PBMCs), 2.8 ± 1.1 in PBMCs, and 4.0 ± 2.1 in liver.
| Fig. 2. HVR 1 amino acid sequence of HCV and numbers of clones obtained from serum, PBMCs, and liver in all patients. Eight to 14 subclones were obtained from each tissue. Identical clones are represented by a combination of a number and a letter such as 1a, 1b, or 1c. The percent amino acid difference of each clone was calculated on the basis of the amino acid sequence of the predominant clone (amino acid sequence in the uppermost line). Amino acid residues are represented by a single-letter code and numbered according to the sequence of HCV-J4.29 Horizontal bars indicate amino acids identical to those of the predominant clone. |
DISCUSSION
A cross-mixing test of PBMCs from healthy controls with HCV-RNA-positive serum showed false detection of HCV RNA in PBMCsas a result of the adsorption of HCV from serum in a control,and also demonstrated that this contamination could be eliminatedby treatment with trypsin-EDTA and subsequently with RNaseA.
Great care must be taken to assess the HCV replication in PBMCs, because the evidence of HCV replication is based on the detectionof negative-strand HCV RNA,3-10 and while specific and sensitivemethods for detecting negative-strand HCV RNA are currently beingdeveloped, they are not yet established.30,31 In the presentstudy as well, there were discrepant results between two methodsfor detecting negative-strand HCV RNA. Although negative-strandHCV RNA in PBMCs was not detected with a highly strand-specificRT-PCR method (method II) in any patient examined, these resultsdo not necessarily exclude the possibility of HCV replicationin PBMCs. The sensitivity of this method appeared to be limited,because negative strands in liver were not detected in 1 of 3 patients. A less specific but more sensitive method (method I)than method II could detect negative-strand HCV RNA in PBMCs in7 of 9 patients. High sensitivity, as well as high specificity,is required for detecting negative strands, especially in cellscontaining a large molar excess of positive strands, because negative-strandHCV RNA can be potentially driven into duplexes with positivestrands. These results indeed reflect the current controversyconcerning the methodology for negative-strand HCV RNA assay andthe need for an alternative method for proving HCV replicationinPBMCs.
Our present study demonstrated differences in the complexity and specific sequences of HVR 1 quasispecies among serum, PBMCs,and liver, which is indicative of infection of PBMCs by HCV. Thesefindings are compatible with the observations that different quasispeciesof HVR 1 existed in serum, PBMCs, and liver in chimpanzees thathad been inoculated with HCV strain H7732 and in patients withchronic hepatitis C.33 In the chimpanzee study, an HCV strainunique to PBMCs was found, but the strain did not persist eitherin PBMCs or in liver 6 years later. This observation suggeststhat HCV strains unique to PBMCs are not always present in vivo,which is consistent with the present findings that an amino acidsequence unique to PBMCs was absent in 5 patients. The observationby Maggi et al.33 that the predominant sequence was the samein serum, PBMCs, and liver in only 1 of 10 patients was not consistentwith our results. This inconsistency may be derived from the differencein methods used, i.e., single-strand conformation polymorphismanalysis versus cloning and sequencing analysis. Because HVR 1 is the most heterogeneous domain in the HCV genome, the possibilityshould be taken into consideration that artificial quasispeciesmay be generated during RT-PCR.34 We previously demonstratedhigh homology as well as considerable differences in HVR 1 quasispeciesbetween immune complexes and nonimmune complexes, using exactlythe same methods as in the present study.28 In addition, anotherfive subclones from each tissue were examined for the HVR 1 quasispeciesin patient 8, who showed the most heterogeneous quasispecies,but the results were almost the same. These results indicatedthat the different complexity of HVR 1 quasispecies found in eachtissue in all patients was unlikely to be the result of artificialsubstitution of the virus sequence. Next, we should consider whetherthe complexity of HVR 1 quasispecies in liver may vary accordingto the site of liver biopsy. Regarding this issue, clusteringof similar clones has been shown to be present in three differentparts of the liver in patients with chronic hepatitis C35; therefore, sampling problems related to liver biopsy are unlikelyto be linked to the different complexity of HVR 1 quasispeciesin eachtissue.
The heterogeneity of HVR 1 quasispecies observed in 3 patients (23%), who also showed differences in the predominant clonein each tissue, might result in or result from different neutralizationwith antibodies to HVR 1, because antibodies to HVR 1 have beenshown to have neutralizing activity and are found in persistentlyinfected patients.18,36,37 The tendency of the complexityof HVR 1 quasispecies to be greater in serum than in PBMCs orliver, and the presence of HCV variants common to serum and PBMCs,suggests that the quasispecies of circulating HCV are derivedfrom HCV in both PBMCs and liver. This also implies the potentialrelease of virions from PBMCs, even though there is no evidenceas yet of the active release of virions from PBMCs. On the otherhand, it was uncertain why certain variants were present in serum,but not in PBMCs or liver, in all patients except patient 9. Theseresults were consistent with those of Maggi et al.,33 who foundvariants unique to serum in 7 of 10 patients. This does not necessarilyimply that serum-specific sequences are derived from yet anothersite, i.e., not liver or PBMCs. This could occur if such variantsrepresent a very small portion of the quasispecies in PBMCs orliver, but are activelyreleased.
A marked difference in the complexity of HVR 1 quasispecies in serum between before and after interferon therapy has beenobserved, possibly indicative of a different viral sensitivityto interferon, selective immune pressure by the host, or both.38,39We and others have shown that persistence of HCV RNA in PBMCsduring interferon therapy, in spite of normalization of the alaninetransaminase level and the disappearance of HCV RNA from serum,is predictive of subsequent flare-ups of the alanine transaminaselevel and the reappearance of HCV RNA in serum after therapy.40,41It is uncertain why HCV RNA persisted in PBMCs in spite of theloss from serum, but HCV variants that are resistant to interferonmay have been selected in PBMCs. Because the complexity of HVR1 quasispecies differs between PBMCs and liver, as demonstratedin the present study, HCV may have different sensitivity to interferonin the two sources. Some minor clones observed in this study thatare unique to PBMCs or common to serum and PBMCs, but not foundin liver, may be linked with poor sensitivity to interferon, thoughfurther studies are required to clarify theseissues.
Acknowledgment
The authors thank Dr. William F. Carman, Glasgow University, Scotland, for his critical reading of the manuscript. They also thank Mr. K. Barrymore, Sapporo Medical University, Sapporo, Japan, for help with the manuscript.
Abbreviations
Abbreviations: HCV, hepatitis C virus; PBMCs, peripheral blood mononuclear cells; RT-PCR, reverse-transcription polymerase chain reaction; HVR 1, hypervariable region 1; EDTA, ethylenediaminetetraacetic acid.
Footnotes
Received January 20, 1998; accepted July 31, 1998.
Address reprint requests to: Keisuke Hino, M.D., First Department of Internal Medicine, Yamaguchi University, School of Medicine, 1144 Kogushi, Ube, Yamaguchi, 755-8505 Japan. E-mail: k.hino@po.cc.yamaguchi-u.ac.jp; fax: (81) 836-22-2240.
REFERENCES
| 1. | Cleaves GR, Ryan TE, Schlesinger RW. Identification and characterization of type 2 Dengue virus replicative intermediate and replicative form RNAs. Virology 1981;111:73-78[Medline]. |
| 2. | Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for Flavivirus gene expression and evolution. Science 1985;229:726-733[Medline]. |
| 3. | Wang JT, Sheu JC, Lin JT, Wang TH, Chen DS. Detection of replicative form hepatitis C virus RNA in peripheral blood mononuclear cells. J Infect Dis 1992;166:1167-1169[Medline]. |
| 4. | Willems M, Peerlinck K, Moshage H, Deleu I, Van den Eynde C, Vermylen J, Yap SH. Hepatitis C virus-RNAs in plasma and in peripheral blood mononuclear cells of hemophiliacs with chronic hepatitis C: evidence for viral replication in peripheral blood mononuclear cells. J Med Virol 1994;42:272-278[Medline]. |
| 5. | Muller HM, Pfaff E, Goeser T, Kallinwski B, Solbach C, Thielman L. Peripheral blood leukocytes serve as a possible extrahepatic site for hepatitis C virus replication. J Gen Virol 1993;74:669-676[Medline]. |
| 6. | Moldvay BJ, Deny P, Pol S, Brechot C, Lamas E. Detection of hepatitis C virus RNA in peripheral blood mononuclear cells of infected patients by in situ hybridization. Blood 1994;83:269-273[Abstract]. |
| 7. | Lerat H, Berby F, Trabaud MA, Vidalin O, Major M, Trepo C, Inchauspe G. Specific detection of hepatitis C virus strand RNA in hematopoietic cells. J Clin Invest 1996;97:845-851[Abstract/Full Text]. |
| 8. | Takehara T, Hayashi N, Mita E, Hagiwara H, Ueda K, Katayama K, Kasahara A, et al. Detection of the minus strand of hepatitis C virus RNA by reverse transcription and polymerase chain reaction. HEPATOLOGY 1992;15:387-390[Abstract]. |
| 9. | McGuinness PH, Bishop GA, McCaughan GW, Trowbridge R, Gowans EJ. False detection of negative-strand hepatitis C virus RNA. Lancet 1994;343:551-552. |
| 10. | Lanford RE, Chavez D, Chisari FV, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol 1995;69:8079-8083[Abstract]. |
| 11. | Mizutani T, Kato N, Saito S, Ikeda K, Sugiyama K, Shimotohno K. Characterization of hepatitis C virus replication in cloned cells obtained from a human T-cell leukemia virus type 1-infected cell line, MT-2. J Virol 1996;70:7219-7223[Abstract]. |
| 12. | Shimizu YK, Iwamoto A, Hijikata M, Purcell RH, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T cell line. Proc Natl Acad Sci U S A 1992;89:5477-5481[Medline]. |
| 13. | Shimizu YK, Purcell RH, Yoshikura H. Correlation between the infectivity of hepatitis C virus in vivo and its infectivity in vitro. Proc Natl Acad Sci U S A 1993;90:6037-6041[Medline]. |
| 14. | Ikeda M, Kato N, Mizutani T, Sugiyama K, Tanaka K, Shimotohno K. Analysis of the cell tropism of HCV by using in vitro HCV-infected human lymphocytes and hepatocytes. J Hepatol 1997;27:445-454[Medline]. |
| 15. | Martell M, Esteban J, Ouer J, Genesca J, Weiner A, Esteban R, Guardia J, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol 1992;66:3225-3229[Abstract]. |
| 16. | Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable region in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun 1991;175:220-228[Medline]. |
| 17. | Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Sekiya H, Hijikata M, Shimotohno K. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem Biophys Res Commun 1992;189:119-127[Medline]. |
| 18. | Kato N, Ootsuyama Y, Sekiya H, Ohkoshi S, Nakazawa T, Hijikata M, Shimotohno K. Genetic drift in hypervariable region 1 of the viral genome in persistent hepatitis C virus infection. J Virol 1994;68:4776-4784[Abstract]. |
| 19. | Kurosaki M, Enomoto N, Marumo F, Sato C. Rapid sequence variation of the hypervariable region of hepatitis C virus during the course of chronic infection. HEPATOLOGY 1993;18:1293-1299[Abstract]. |
| 20. | Lawal Z, Petric J, Wong VS, Alexander GJA, Allain JP. Hepatitis C virus genomic variability in untreated and immunosuppressed patients. Virology 1997;228:107-111[Medline]. |
| 21. | Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis: diagnosis, grading and staging. HEPATOLOGY 1994;19:1513-1520[Medline]. |
| 22. | Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, et al. Typing hepatitis C virus by polymerase chain with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol 1992;73:673-679[Medline]. |
| 23. | Hino K, Okuda M, Konishi T, Ishiko H, Okita K. Serial assay of hepatitis C virus RNA in serum for predicting response to interferon- |
| 24. | Hino K, Fujii K, Korenaga M, Murakami C, Okazaki M, Okuda M, Okita K. Correlation between relative number of circulating low-density hepatitis C virus particles and disease activity in patients with chronic hepatitis C. Dig Dis Sci 1997;42:2476-2481[Medline]. |
| 25. | Okamoto H, Okada S, Sugiyama Y, Tanaka T, Sugai Y, Akahane Y, Machida A, et al. Detection of hepatitis C virus RNA by a two-stage polymerase chain reaction with two pairs of primers deduced from the 5′-noncoding region. Japan J Exp Med 1990;60:215-222[Medline]. |
| 26. | Fujii K, Hino K, Okazaki M, Okuda M, Kondoh S, Okita K. Differences in hypervariable region 1 quasispecies of hepatitis C virus between human serum and peripheral blood mononuclear cells. Biochem Biophys Res Commun 1996;225:771-776[Medline]. |
| 27. | Kwok S, Higuchi R. Avoiding false positives with PCR. Nature 1989;339:237-238[Medline]. |
| 28. | Korenaga M, Hino K, Okazaki M, Okuda M, Okita K. Differences in hypervariable region 1 quasispecies between immune complexed and non-immune complexed hepatitis C virus particles. Biochem Biophys Res Commun 1997;240:677-682[Medline]. |
| 29. | Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore EE, et al. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 1992;190:894-899[Medline]. |
| 30. | Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand specific RT/PCR. Virology 1994;202:606-614[Medline]. |
| 31. | Gunji T, Kato N, Hijikata M, Hayashi K, Saitoh S, Shimotohno K. Specific detection of positive and negative stranded hepatitis C viral RNA using chemical RNA modification. Arch Virol 1994;134:293-302[Medline]. |
| 32. | Shimizu YK, Igarashi H, Kanematsu T, Fujiwara K, Wong DC, Purcell RH, Yoshikura H. Sequence analysis of the hepatitis C virus genome recovered from serum, liver, and peripheral blood mononuclear cells of infected chimpanzees. J Virol 1997;71:5769-5773[Abstract]. |
| 33. | Maggi F, Fornai C, Linda M, Giorgi M, Morrica A, Pistello M, Cammarota G, et al. Differences in hepatitis C virus quasispecies composition between liver, peripheral blood mononuclear cells and plasma. J Gen Virol 1997;78:1521-1525[Medline]. |
| 34. | Smith DB, McAllister J, Casino C, Simmonds P. Virus `quasispecies’: making a mountain out of a molehill? J Gen Virol 1997;78:1511-1519[Medline]. |
| 35. | Sakai A, Honda M, Matsushita E, Kaneko S, Kobayashi K. Hepatitis C virus quasispecies in three different parts of a liver and serum in patients with chronic hepatitis C [Abstract]. HEPATOLOGY 1997;26:300A. |
| 36. | Shimizu YK, Hijikata M, Iwamoto A, Alter HJ, Purcell RH, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol 1994;68:1494-1500[Abstract]. |
| 37. | Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, et al. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A 1994;91:7792-7796[Medline]. |
| 38. | Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. HEPATOLOGY 1992;16:619-624[Abstract]. |
| 39. | Moribe T, Hayashi N, Kanazawa Y, Mita E, Fusamoto H, Negi M, et al. Hepatitis C virus complexity detected by single-strand conformation polymorphism and response to interferon therapy. Gastroenterology 1995;108:789-795[Abstract]. |
| 40. | Saleh MG, Tibbs CJ, Koskinas J, Pereira LMMB, Bomford AB, Portmann BC, McFarlane IG, et al. Hepatic and extrahepatic hepatitis C virus replication in relation to response to interferon therapy. HEPATOLOGY 1994;20:1399-1404[Abstract]. |
| 41. | Okuda M. HCV-RNA assay in peripheral blood mononuclear cells in relation to IFN therapy. Gastroenterol Jpn 1993;28:535-540[Medline]. |
Copyright © 1999 by the American Association for the Study of Liver Diseases.