Full-Length Complementary DNA of Hepatitis C Virus Genome From an Infectious Blood Sample
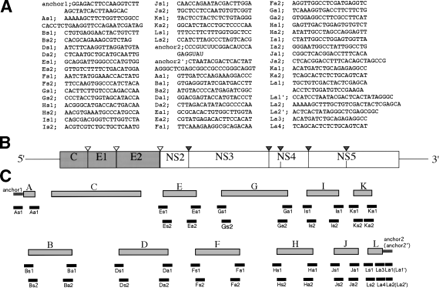
Fig. 2. Construction of a full-length cDNA clone of HCV. (A) Sequences of primers and anchors for RT-nested PCR. The synthetic oligonucleotide primers are according to the published nucleotide sequences of type Ib HCV clones and a novel 98-nt sequence in the 3′ terminus. The anchors (anchor 1, anchor 2′) and the anchors’ primers (As1, La1′, La2′) are obtained from the 5′-Ampli FINDER RACE kit and Marathon cDNA amplification kit. An oligo RNA (anchor 2′) and primers (Ls1, Ls2, Ls3, Ls4, La1, La2) were according to Tanaka et al.63 (B) Gene organization of the HCV. Shaded and open boxes indicate structural and nonstructural proteins, respectively. UTRs are indicated by bars. Black and white triangles indicate cleavage sites by cellular signalase and viral-coded proteases, respectively. (C) Strategy for making a full-length cDNA clone of HCV. The locations of primers and anchors are shown. The 5′ and 3′ terminal sequences were determined by use of a 5′-Ampli FINDER RACE kit and an RNA linker ligation, respectively.