Full-Length Complementary DNA of Hepatitis C Virus Genome From an Infectious Blood Sample
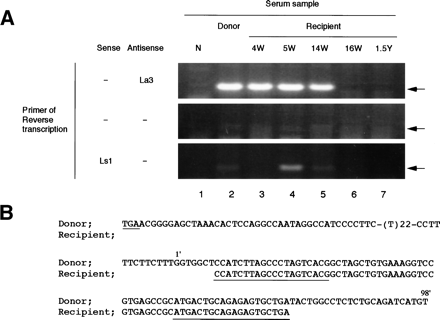
Fig. 3. Detecting and sequencing of the 3′ terminal sequence of HCV from donor and recipient plasma. (A) Detection of the terminal extra sequence in positive and negative strands of HCV RNA and comparison with the presence of 5′ UTR sequence. The presence of the 3′ extra sequence was examined by RT-nested PCR and electrophoresed on a 3% agarose gel as described in Materials and Methods. The primers used in RT are indicated at the left. Arrows indicate the expected size of RT-PCR products. Lane 1, normal serum (N); lane 2, plasma sample of the donor; lane 3, recipient sample 4 weeks posttransfusion (4W);lane 4, 5 weeks posttransfusion (5W); lane 5, 14 weeks posttransfusion (14W); lane 6, 16 weeks posttransfusion (16W); and lane 7, 1.5 years posttransfusion (1.5Y). (B) Homology analysis of the 3′ terminal sequence of HCV derived from donor and recipient. The upper sequences represent the 3′ terminal sequence of pNIHJ1. The lower sequences indicate the terminal sequences obtained from the recipient 4 weeks posttransfusion. The nucleotides in the 3′ extra sequences are numbered 1′ to 98′ above the sequences. The termination codon of the ORF (TGA) and sequences of the primers used in PCR are underlined.