HEPATOLOGY, March 1998, p. 877-880, Vol. 27, No. 3
Original Articles
Quantitation of Hepatitis G and C Viruses in the Liver: Evidence That Hepatitis G Virus Is Not Hepatotropic
Mario G. Pessoa1,2, Norah A. Terrault1,2, Jill Detmer3, Janice Kolberg3, Mark Collins3, Howayda M. Hassoba1,2, and Teresa L. Wright1,2
From the 1 Departments of Medicine, University of California, San Francisco, CA;2 Department of Veterans Affairs Medical Center and 3 Chiron Corporation, Emeryville, CA
ABSTRACT
Hepatitis G virus (HGV) is prevalent in patients with chronic liver disease and has been previously detected in liver specimens. However, it is unknown whether the virus is replicating in the liver or is simply a contaminant from serum. We sought to determine whether HGV was hepatotropic and to determine whether coinfection with HGV and Hepatitis C virus (HCV) influenced the level of either virus. Virus was quantitated using branched DNA (bDNA) assay for both HGV and HCV in the liver explants and pre-transplant serum samples from 30 transplant recipients: Group I, HGV/HCV coinfection (n = 10); group II, HCV infection alone, (n = 8); group III, HGV alone (n = 12). In patients with coinfection HCV (RNA) titers in liver were consistently higher than those for HGV RNA (median 1.13 × 108 and 360,000 Eq/g respectively, P < .01). The ratio of liver/serum viral RNA was significantly higher for HCV than for HGV (median 129 and 0.3 respectively, P < .01). Levels of HCV RNA were similar in patients with HCV infection alone versus those with HGV/HCV coinfection (median; liver = 1.15 × 107 vs. 1.13 × 108 Eq/g, serum = 500,000 vs. 200,000 Eq/mL) and levels of HGV RNA in liver and serum were similar in patients with HGV infection alone compared to those with HGV/HCV coinfection (median; liver = 1.2 × 106 vs. 4.0 × 105 Eq/g, serum = 4.5 × 106 vs. 2.6 × 106 Eq/mL). Levels of either virus appeared unaffected by the presence of an additional virus. The high ratio of HCV RNA levels in liver compared to serum is consistent with its known hepatotropism, but this pattern was not observed for HGV. The median liver/serum ratio of HGV RNA was less than unity, a finding consistent with serum contamination of liver tissue. Thus we conclude that the liver is not the main site of HGV replication. (HEPATOLOGY 1998;27:877-880).
INTRODUCTION
Hepatitis G virus (HGV) is a positive strand (RNA) virus of the Flaviviridae family whose transmissibility and persistence of viremia has been established by animal and human studies, 1,2 but whose clinical significance as a hepatitis virus has become increasingly controversial. Multiple studies have been published showing no association between HGV and either acute or chronic liver disease.3-6 If HGV was a hepatotropic virus with replication within liver cells, one would expect to detect HGV RNA in liver tissue. If HGV behaved like Hepatitis C virus (HCV), a known hepatotropic virus, the ratio of HGV RNA in liver and serum would be predicted to be similar to that found for HCV. In patients with chronic HCV infection, the level of HCV RNA measured in liver by branched DNA (bDNA) assay, ranges from 5.3 × 106 to 1.35 × 109 Eq/g, with a geometric mean value of 3.7 × 107 to 1.40 × 108 Eq/g.7-9 One study demonstrated that the levels of HCV RNA in liver tissue varied with severity of liver disease in a heterogeneous population with chronic persistent hepatitis, chronic active hepatitis, and cirrhosis,7 but in all cases the levels of HCV RNA were much higher in liver than in serum. The correlation between liver and serum HCV RNA levels has been examined by several investigators, but no single numerical constant has been defined. The mean ratio of HCV RNA in liver versus serum varies from 36 to 130 Eq per gram/Eq per mL.7-11
In this study we compared the amount of HGV RNA with HCV RNA in liver and serum from patients with end-stage liver disease. We hypothesized that if HGV were a hepatotropic virus, behaving in a manner similar to HCV, levels of HGV RNA in liver would be higher than those in serum. As a secondary aim, we sought to determine whether coinfection of HGV and HCV suppressed the replication of either virus individually by comparing viral levels in three groups of patients; those with coinfection; those with HCV infection alone; and those with HGV infection alone.
PATIENTS AND METHODS
Study Population. Thirty patients undergoing liver transplantation at the University of California at San Francisco between February, 1988, and December, 1995, were divided into three groups as follows:
Study Group. Group I consisted of ten patients with HCV-related end-stage liver disease and HGV co-infection prior to liver transplantation. HCV infection was defined by the presence of detectable anti-HCV by second generation enzyme-linked immunoassay and detectable HCV RNA by polymerase chain reaction (PCR). HGV infection was defined by the presence of detectable HGV RNA by PCR. HCV RNA was measured in the same samples as HGV RNA and thus served as a positive control for RNA preservation.
Control Groups. Group II consisted of eight patients with HCV-related end-stage liver disease, without HGV co-infection prior to liver transplantation. Group III consisted of 12 patients with HGV infection prior to liver transplantation, undergoing liver transplantation for end-stage liver disease of non-viral etiologies. The diagnoses in group III included the following: 1) cryptogenic cirrhosis (n = 3); 2) alcoholic liver disease (n = 3); 3) primary biliary cirrhosis (n = 2); 4) primary sclerosing cholangitis (n = 2); 5) autoimmune hepatitis (n = 1); and 6) Budd-Chiari syndrome (n = 1). All patients were negative for Hepatitis B surface antigen, antibody to Hepatitis B core, and HCV RNA prior to liver transplantation. All patients gave consent under an Institutional Review Board approved protocol.
Sample Collection and Processing. Serum samples and liver tissue (explants) were collected on the same day. Liver specimens were processed using a modification of the procedure described by Cox,12 and previously reported by our group.9
Virological Methods
HGV Detection. Branched-DNA assay: HGV RNA was quantitated by bDNA assay in pre-transplant serum samples (Eq/mL) and liver tissue (Eq/g) of all patients, using a research based bDNA assay as described previously.6 Results were expressed as HGV RNA Eq per mL for serum specimens, and HGV RNA Eq per gram wet weight for liver tissue. One equivalent was defined as the amount of HGV RNA that generated a level of light emission equivalent to that generated by one copy of quality level 1 RNA standard.13 The HGV bDNA assay had a provisional limit of detection of approximately 75,000 Eq/mL and 300,000 Eq/g, based on dilutions of a 700 nucleotide synthetic HGV RNA transcript. All bDNA tests were performed at Chiron Corporation (Emeryville, CA).
Reverse Transcriptase-PCR. Presence of HGV RNA in serum was determined by reverse transcriptase-PCR amplification assay, as previously described.14
HCV Detection. Branched-DNA Assay. HCV RNA was quantitated in both pre-transplant serum samples and liver tissue of all patients by bDNA assay (Quantiplex® 2.0, Chiron Corporation, Emeryville, CA). Results were expressed as HCV RNA Eq per mL for serum specimens and HCV RNA Eq per gram wet weight for liver tissue. The quantitation limit of the Quantiplex® HCV RNA 2.0 assay is 200,000 Eq per mL.15 All runs included both positive and negative controls for serum or liver tissue.
RT-PCR assay: Serum samples which were negative for HCV RNA by bDNA assay were re-tested, using a RT-PCR amplification assay as previously described.16
Statistical Analysis
The Wilcoxon matched pairs test was used to compare levels of the different viruses in the same liver and serum samples. The Mann Whitney U test was used to compare viral RNA values between patient groups. For statistical purposes, patients that were HGV RNA and/or HCV RNA negative by bDNA assay, but were positive by PCR, were assigned with half of the value of the bDNA assay (100,000 Eq/mL for HCV RNA and 37,500 Eq/mL for HGV RNA).
RESULTS
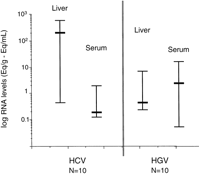
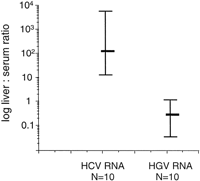
Viral Levels in Study Group. HCV RNA levels were significantly higher in liver than serum (P < 0.01). HGV RNA levels were significantly lower in liver than serum (P = .03) (Fig. 1). Levels of HCV RNA in liver were consistently higher than levels of HGV RNA in liver (median; 1.13 × 108 Eq/g vs. 3.6 × 105 Eq/g, respectively; P < .01). Levels of HCV RNA in serum were lower than levels of HGV RNA in serum (median; 1.7 × 105 Eq/mL vs. 2.6 × 106 Eq/mL, respectively;P = .01) (Fig. 1). The median ratio of liver/serum HCV RNA in coinfected patients was 129 while the median ratio for HGV RNA was 0.3; P < .01 (Fig. 2).
|
|
Fig. 1. Quantitation of HCV and HGV viral RNA in liver and serum in patients with HGV/HCV coinfection (study group). HCV RNA levels were significantly higher in liver than serum (P< .01; Wilcoxon matched pairs test). HGV RNA levels were significantly lower in liver than serum (P = .03; Wilcoxon matched pairs test). Hepatic HCV RNA levels were significantly higher than those for HGV RNA (P < .01; Wilcoxon matched pairs test). |
|
|
Fig. 2. HCV and HGV liver/serum ratios in patients with HGV/HCV coinfection (study group). Liver/serum ratios were significantly higher for HCV RNA compared with HGV RNA (P < .01; Wilcoxon matched pairs test). |
Viral Levels in All Patient Groups. The median ratio of HCV RNA in liver/serum in all patients with HCV infection (n = 18) was significantly higher than the overall median ratio of HGV RNA in all patients with HGV infection (n = 22) (113 and 0.3 respectively; P < .01).
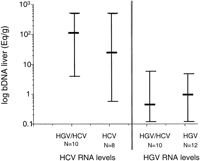
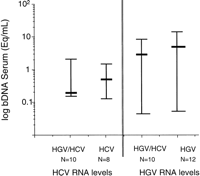
Interaction of Both Viruses. Levels of HCV RNA in liver were similar in HCV infected patients alone compared to HGV/HCV coinfected patients (median; 1.15 × 107 Eq/g vs. 1.13 × 108 Eq/g, respectively; P = .29) and levels of HGV RNA in liver were similar in patients with HGV infection alone compared with those with HGV/HCV coinfection (median; 1.2 × 106 Eq/g vs. 4 × 105 Eq/g, respectively; P = .33) (Fig. 3). Similarly, viral RNA levels in serum were not significantly different in patients infected with only one virus compared with those with coinfection (HCV = 4.6 × 105 vs. 1.7 × 105 Eq/mL, respectively; P= 0.96 and HGV = 4.5 × 106 vs. 2.6 × 106 Eq/mL, respectively; P = .23) (Fig. 4).
|
|
Fig. 3. Effect of HGV/HCV coinfection on hepatic viral levels of HCV or HGV. Levels of HCV RNA in liver were similar in patients with HCV infection alone compared with those with HGV/HCV coinfection (P = .29; Mann Whitney U test); levels of HGV RNA in liver were similar in patients with HGV infection alone compared with those with coinfection (P = .33; Mann Whitney U test). |
|
|
Fig. 4. Effect of HGV/HCV coinfection on serum viral levels of HCV or HGV. Levels of HCV RNA in serum were similar in patients with HCV infection alone compared with those with HGV/HCV coinfection (P = .96; Mann Whitney U test); levels of HGV RNA in serum were similar in patients with HGV infection alone compared with those with coinfection (P= .23; Mann Whitney U test). |
DISCUSSION
The liver is the major site of viral replication for HCV. Quantitation of virus in liver tissue and comparison with levels of virus in serum is an indirect measure of hepatotropism. Several studies had quantitated HCV RNA in liver, with a broad range of geometric mean titers between different studies.7-11 This difference may be due to variability in the stage of liver disease and duration of infection or due to methodological differences in viral quantitation.7 Liver HCV RNA levels were not shown to be related to severity of histological activity when a homogeneous group of patients with chronic hepatitis was assessed.8 However, there is general agreement that HCV RNA levels in liver are greater than HCV RNA levels in serum by at least a one log range. In this study, we used liver specimens from a homogeneous group of patients with end-stage liver disease undergoing liver transplantation to evaluate further the hepatotropism of known HCV and possible HGV hepatotropic viruses.
In the study group with HGV/HCV coinfection, median levels of HCV RNA were significantly higher in liver compared to serum, consistent with the known tropism of this virus for the liver. This was not observed in patients with HGV; the median levels of HGV RNA were lower in liver than in serum. Calculation of liver/serum ratios of viral RNA depicted more clearly the difference between the two viruses. The median liver/serum ratio of HCV RNA was greater than 100, while the median liver/serum ratio of HGV RNA was less than 1. These findings were also true when liver/serum ratios were calculated in the study population as a whole, comparing patients with HCV and HGV/HCV co-infection to those with HGV and HGV/HCV coinfection. If HGV were replicating in the liver, the liver/serum ratios of HGV RNA might be predicted to be comparable to that of HCV RNA. Our findings suggest HGV is not replicating in the liver, however, alternative interpretations exist. The low liver/serum ratio of HGV RNA in HGV infected patients may indicate that HGV replicates at a very low rate, or that replication within the hepatocytes is followed by rapid export of the virus into the serum. In a recent study from Madejon et al., both genomic and antigenomic strands of GBV-C RNA were identified in liver.17 If true, this information would suggest that HGV replication is occurring in the liver but at a very low rate. However, given the methodological problems in quantitating accurately the negative strand of GBV-C/HGV, confirmation of these results must be awaited before concluding that HGV is an hepatropic virus. Alternatively, our data can potentially be explained by differences in the clearance of HCV and HGV from the circulation. Thus, even if both viruses were produced exclusively in the liver, a higher liver/serum ratio could be observed for HCV if it were cleared more rapidly from the serum than HGV. While further study of the dynamic relationship of these viruses between compartments is required, our findings support the weak hepatotropism of HGV.
There is evidence of interaction between known hepatotropic viruses. HCV exerts a suppressive effect in Hepatitis B virus (HBV) and may enhance the clearance of HBV antigens.18 HBV can also suppress HCV replication, but this effect is less marked than the suppressive effect of HCV on HBV.18 In our study, levels of HCV RNA in liver were similar in patients with HCV infection alone compared to patients with HGV/HCV coinfection. The converse was also true. Levels of HGV RNA in liver were comparable in patients with HGV infection alone and patients with HGV/HCV coinfection. This pattern was reproduced in serum, showing that levels of either virus were unaffected by the presence of an additional virus. Thus in our study, no interactions between HGV and HCV could be shown.
In conclusion, our findings raise serious doubts about the hepatotropic nature of HGV and support epidemiological studies which have failed to show an association between HGV and liver disease.
ADDENDUM
After submission of this work, a letter to the editor was published on the same subject.19
Footnotes
Abbreviations: bDNA, branched DNA; HBV, Hepatitis B virus; HCV, Hepatitis C virus; HGV, Hepatitis G virus; PCR, polymerase chain reaction; RNA, ribonucleic acid.
TLW is supported by National Institutes of Health grant R29AI32242, a Veterans Administration Merit Review, and by NIDDK grant DK-26743 (Liver Center). NAT is supported by the International Hepatitis Foundation
Received July 15, 1997; accepted November 20, 1997.
Address reprint requests to: Teresa L. Wright, M.D., Gastroenterology Unit, IIIB, Veterans Affairs Medical Center, 4150 Clement St., San Francisco, CA 94121. Fax: (415) 750-2196.
Copyright © 1998 by the American Association for the Study of Liver Diseases.