Effects of Chronic Ethanol Administration on Rat Liver Proteasome Activities: Relationship With Oxidative Stress
Hepatology, January 1999, p. 14-20, Vol. 29, No. 1
Virginie Fataccioli1,
From the 1Laboratory of Biomedical Research on Alcoholism, Univ. René Descartes, Paris, France; and 2Department of Pathology, HARBOR-UCLA Medical Center, Torrance, CA.
ABSTRACT
We previously reported that ethanol elicits an increased protein oxidation in the liver of rats receiving chronic ethanol by continuous intragastric infusion (Tsukamoto-French method). This accumulation of oxidized proteins could result from a decrease in the cytosolic proteolysis, related specifically to alkaline protease and its major components, the proteasomes. Because several studies suggest that intracellular proteolysis depends on the severity of oxidative stress, we investigated the cytosolic proteolytic activity under two chronic ethanol treatment paradigms associated with varying degrees of oxidative stress. For 4 weeks, male rats received chronic ethanol by continuous intragastric infusion or by oral administration (10% ethanol ad libitum as sole drinkingfluid). A significant decrease was evident for alkaline proteaseactivity as well as for sodium dodecyl sulfate (SDS)-activatedlatent 20S proteasome (chymotrypsine-like [ChT-L] and peptidylglutamyl peptide hydrolase [PGPH] activities) in the liver of rats receiving ethanol by continuous intragastric infusion. Free radical production and related processes appeared to be contributing events in proteolysis inhibition, because phenethyl isothiocyanate (PIC), an inhibitor of cytochrome P4502E1 (CYP2E1), reduced the inhibition of the ethanol-related ChT-L activity. Moreover, the lipid peroxidation level was inversely correlated with ChT-L activity. In contrast, no such changes were observed in ChT-L and PGPH activities or in cellular free radical targets following the oral ad libitum consumption of 10% ethanol. It appears, thus, that only the alcohol treatment paradigm associated with an overt oxidative stress produced a significant inhibition of the proteasome activity. The mechanisms of proteasome inhibition could involve the formation of an endogenous inhibitor such as protein aggregates or aldehyde-derivative peptides. Whatever the mechanism, the inhibition of cytosolic proteolysis and the subsequent accumulation of damaged proteins may be involved in the oxidatively challenged alcoholic livers and play a pathogenic role in experimental alcoholic liver disease. (HEPATOLOGY 1999;29:14-20.)
INTRODUCTION
There are two separate mechanisms that may be involved in intracellular protein breakdown: lysosomal and extralysosomal. The lysosomal pathway is the general mechanism involved in long-lived protein degradation. The main cytoplasmic proteases identified so far include the calpains, calcium-dependent proteases,1 and two large degradative enzymes, the multicatalytic protease (MCP) or 20S proteasome and the 26S proteasome, 2-4 major components of the alkaline proteases. The ubiquitin-dependent pathway, related to the 26S proteasome, plays an important role in the removal of abnormal or short-lived regulatory proteins,4 whereas the 20S proteasome appears to be more likely implicated in the removal of abnormal and oxidized proteins,5-7 in a ubiquitin-independent pathway. The wide distribution of the proteasomes in eukaryoticcells8 has led to the suggestion that they may serve a fundamentally important biological role. They may also constitute a “secondary antioxidant defense system”9 that protects critical regulatorymolecules from permanent oxidative damage. In fact, it has beenshown that oxidative damage of proteins increases their susceptibility to proteolysis10 as long as extensive modifications of the putative substrates are not induced.
The 20S proteasome expresses at least three distinct peptidase activities, i.e., chymotrypsin-like (ChT-L), trypsin-like (T-L), and peptidylglutamyl peptide hydrolase (PGPH) activities.11. In addition, the complex has been shown to exist in a latent form that can be specifically activated by agents such as sodium dodecyl sulfate (SDS).11,12 Ethanol could induce alterations in hepatic protein turnover, predominantly a decrease in the rate of protein degradation.13,14 Recent studies have indicated that decreased hepatic lysosomal proteolysis is a primary factor responsible for ethanol-induced hepatic protein accumulation.15,16It has been also reported that ethanol administration, using theLieber-DeCarli model, elicited a rise in hepatocellular ubiquitinlevel, a crucial component of the extralysosomal proteolytic pathway.17 Whether such an accumulation of ubiquitin and ubiquitin protein conjugates results from a decrease in the proteasome pathway has not yet been investigated. Moreover, long-term ethanol consumption is associated with varying degrees of oxidative stress, according to the method of ethanol administration. Overt oxidative stress may alter behavior of the substrates and/or enzymes involved in the cytosolic proteolyticpathways.
In the present study, the relationship between ethanol-induced oxidative stress and proteasome activities was examined in rats receiving chronic ethanol by continuous intragastric infusion or by oral administration (10% ethanol ad libitum as the sole drinking fluid). This latter model is characterized by the absence of histologically assessed liver damage18,19 and by similar weight gains and caloric intakes for both controls and treated animals, and it represents “a reasonable model for studies of the effects of moderate alcohol consumption on specific biochemical pathways.”19 The intragastric infusion model20 produces evidence of overt hepatic oxidative stress21,22 associated with histological liver damage,23 including fat accumulation, spotty necrosis, inflammation, and focal fibrosis. Whereas the effect of ethanol on proteasomal activities has been studied on rats fed with ethanol intragastrically for 2 months,24 we examined here both alkaline protease and proteasomal peptidase activities in rats receiving ethanol intragastrically for 1 month, a time when early pathological changes are observed. We also investigated whether the modulation of cytochrome P4502E1 (CYP2E1) expression by phenethyl isothiocyanate(PIC)23 might interfere with ethanol-related changes in proteasome peptidase activities. A preliminary account of this work has been published in an abstract.25
MATERIALS AND METHODS
Chemicals
All chemicals (analytical grade) were purchased from Sigma Chemicals Co. (Paris, France). Protein reagent was from Bio-Rad (Ivry sur Seine, France). The ECL detection kit was from Amersham, Life Science (Les Ulis, France).
Animals and Diet
Intragastric Ethanol Infusion. Male Wistar rats (230-250 g body weight) were purchased from Charles River (Hollister, CA). The permanent intragastric cannula was implanted under sodium thiopental ketamine anesthesia as previouslyreported.20 The rats were maintained according to the Guidelines of Animal Care described by the National Academy of Sciences and published by the National Institutes ofHealth.
The animals were separated into four dietary groups as follows: ethanol (EtOH); isocaloric dextrose (control) pair-fed with ethanol; ethanol and PIC (1 mmol/kg body weight/d) (EtOH + PIC);isocaloric dextrose plus PIC (control + PIC) pair-fed with ethanol + PIC. Rats were intragastrically infused with a high-fat diet plus ethanol or an isocaloric amount of dextrose, with or without PIC, as previously described.26 In this diet, corn oil contributed to 30% to 40% of the total calories. Infusion of the diet and ethanol was continued for 4 weeks. The amount of ethanol was started at 8 g/kg/d and gradually increased to 14 g/kg/d as tolerance developed, such that ethanol represents 24% and 36% of total calories, respectively. Blood alcohol levels were maintained between 200 mg/dL and 300 mg/dL as monitored by the saliva alcohol test (Enzymatic Inc., Horsham, PA). The amount of liquid diet and ethanol were adjusted according to the body weights determined weekly. Other reports using some of the rat liver tissue used in the present study have been published.22,23,26
Oral Ethanol Administration. Male Sprague-Dawley rats were maintained on a standard laboratory diet (Iffa-Credo, U.A.R., Lyon, France). As described previously,27 chronic ethanol-treated rats weighing ![]() 100 g at the start of the experiment were fed the basal diet ad libitum and received an aqueous ethanol solution (10%, vol/vol) ad libitum as sole drinking fluid. The average ethanol intake was 6 to 9 g/kg/d; the ethanol represented approximately 18% of the total energy intake. Control rats were given tap water as fluid. The growth rate of the rats fed ethanol ad libitum (8 g/d) did not differ significantly from that of control animals. At the end of the experimental period (4 weeks), the rats fasted for 16 hours, but the access to the ethanol drinking fluid was maintained to prevent any possible withdrawal stress. The blood ethanol level measured was less than36 ± 2.3 mg/dL.
100 g at the start of the experiment were fed the basal diet ad libitum and received an aqueous ethanol solution (10%, vol/vol) ad libitum as sole drinking fluid. The average ethanol intake was 6 to 9 g/kg/d; the ethanol represented approximately 18% of the total energy intake. Control rats were given tap water as fluid. The growth rate of the rats fed ethanol ad libitum (8 g/d) did not differ significantly from that of control animals. At the end of the experimental period (4 weeks), the rats fasted for 16 hours, but the access to the ethanol drinking fluid was maintained to prevent any possible withdrawal stress. The blood ethanol level measured was less than36 ± 2.3 mg/dL.
Homogenization and Subcellular Fractionation
Liver samples (10% wt/vol) were homogenized either in a solution containing sucrose (0.33 mol/L; pH 7.4) for the determination of alkaline protease activity or in Tris-HCl (50 mmol/L; pH 8.0) containing ethylenediaminetetraacetic acid (0.1 mmol/L) and 2![]() -mercaptoethanol(1 mmol/L) for the determination of peptidase activities, or inHEPES (10 mmol/L; pH 7.4) containing NaCl (137 mmol/L), KCl (4.6 mmol/L), KH2PO4 (1.1 mmol/L), MgSO4 (0.6 mmol/L), leupeptin (0.5 µg/mL), pepstatin (0.7 µg/mL), phenyl methyl sulfonyl fluoride (40 µg/mL), aprotinin (0.5 µg/mL), and ethylenediaminetetraacetic acid (1.1 mmol/L) for the determination of protein carbonyl contentand glutamine synthetase activity.
-mercaptoethanol(1 mmol/L) for the determination of peptidase activities, or inHEPES (10 mmol/L; pH 7.4) containing NaCl (137 mmol/L), KCl (4.6 mmol/L), KH2PO4 (1.1 mmol/L), MgSO4 (0.6 mmol/L), leupeptin (0.5 µg/mL), pepstatin (0.7 µg/mL), phenyl methyl sulfonyl fluoride (40 µg/mL), aprotinin (0.5 µg/mL), and ethylenediaminetetraacetic acid (1.1 mmol/L) for the determination of protein carbonyl contentand glutamine synthetase activity.
The total liver homogenate was centrifuged at 100,000g for 1 hour, and the supernatants (cytosolic fractions) were used for the determination of protease alkaline and peptidase activities. The supernatant used for the determination of protein carbonyl content and glutamine synthetase activity was recovered after centrifugation of liver homogenates at 100,000g for 5 minutes at 4°C.
Assay of Alkaline Protease Activity
Protease activity was assayed by measurement of fluorescamine-reactive products released from intracellular proteins. The 100,000g cytosolic supernatant was extensively dialyzed against a membrane with a molecular-weight cut-off of 12 kd to remove small molecules as amino acids. The dialysis buffer contained Tris-HCl (20 mmol/L; pH 7), magnesium acetate (1 mmol/L), KCl(20 mmol/L), DL-dithiothreitol (0.5 mmol/L), and glycerol (10% vol/vol). Aliquots of the resulting material representing a source of protease and protein substrates were used immediately for protease determination. Typically, proteolysis28 was determined by incubating cytosolic fractions (500 µg to 1 mg of protein) at 37°C during 30 minutes in a reaction mixture (200 µL of final volume) containing HEPES (50 mmol/L; pH 8), KCl (100 mmol/L), MgCl2 (1 mmol/L), and DL-dithiothreitol (5 mmol/L). Reactions were stopped by adding 300 µL 10% trichloroacetic acid, followed by centrifugation at 11,000g for 3 minutes. Next, 200-µL aliquots of trichloroacetic acid-soluble products were added to 2.12 mL of phosphate buffer (0.2 mol/L; pH 8); the final pH was adjusted to 8.0 to 8.2 with NaOH (0.5 N). Each assay was performed at two protein concentrationsat least in duplicate. Free amino groups were then quantifiedwith 800 µL of fluorescamine dissolved in acetone (0.3 mg/mL) as described by Böhlen et al.29 A calibration curve was performedwith glycine.
Assay of Peptidase Activities
ChT-L and PGPH activities were determined with fluorogenic synthetic peptides as N-Succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin(LLVY-AMC) and N-Benzyloxy-carbonyl-Leu-Leu-Glu-2![]() -naphthylamide (LLE-2NA), respectively. Typical assays12 (final volume: 200 µL) contained cytosolic fractions (75 µg of protein), fluorogenic substrates (40 mmol/L), and SDS 0.06% in Tris-HCl (150 mmol/L; pH 8.0). Fluorogenic substrates were dissolved in dimethyl sulfoxide (<4%). The mixture was incubated for 30 minutes at 37°C. The reaction was stopped by adding 1 mL SDS 1% and 2 mL sodium borate (0.1 mol/L; pH 9.1). The peptidase activity was determined fluorometrically by measuring the release of 7-amino-4-methylcoumarin (
-naphthylamide (LLE-2NA), respectively. Typical assays12 (final volume: 200 µL) contained cytosolic fractions (75 µg of protein), fluorogenic substrates (40 mmol/L), and SDS 0.06% in Tris-HCl (150 mmol/L; pH 8.0). Fluorogenic substrates were dissolved in dimethyl sulfoxide (<4%). The mixture was incubated for 30 minutes at 37°C. The reaction was stopped by adding 1 mL SDS 1% and 2 mL sodium borate (0.1 mol/L; pH 9.1). The peptidase activity was determined fluorometrically by measuring the release of 7-amino-4-methylcoumarin (![]() exc = 370 nm,
exc = 370 nm, ![]() em = 430 nm) and 2-naphtylamine (
em = 430 nm) and 2-naphtylamine (![]() exc = 323 nm,
exc = 323 nm, ![]() em = 400 nm) in a Kontron fluorometer.
em = 400 nm) in a Kontron fluorometer.
A standard curve of fluorescence for 7-amino-4-methylcoumarin and 2-naphtylamine was used to calculate the concentration of liberated products in the assays.
In some experiments, the cytosolic supernatant was dialyzed for 18 hours at 4°C against the buffer used for homogenization; the nondialyzed fractions were stored in this buffer at 4°C.30 Cytosolic fractions depleted in MCP were obtained following centrifugation at 100,000g for 6 hours as described by Hedge et al.31
SDS Activation
SDS activation curves were established by measuring peptidase activities in the presence of increasing concentrations of SDS ranging from 0% to 0.1% using 0.005% increments. The maximal activation of ChT-L and PGPH activities was obtained as a plateau between 0.06% and 0.07% SDS.
Western Blots
Purified rat MCP and rabbit immunoserum raised against rat MCP were a kind gift from Dr. B. Friguet.32 The 100,000g supernatant protein (50 µg-75 µg) or purified MCP were subjected to SDS-polyacrylamide gel electrophoresis, performed on a 12% separating gel. Proteins were transferred from the SDS-polyacrylamide gel onto Hybond nitrocellulosemembrane (Amersham) for 1 hour at room temperature in Tris-HCl(25 mmol/L; pH 8.3), glycine (190 mmol/L), and methanol (20% vol/vol). Membrane-bound polypeptides were immunodetected using the ECL (Enhanced ChemiLuminescence) Western blotting analysis system from Amersham.
Other Biochemical Analyses
The assay for measuring lipid peroxide levels in tissue homogenates was performed according to the method described by Uchiyama and Mihara.33 The results were expressed as nanomoles of malondialdehyde (MDA) per milligram of protein. Protein carbonyls were determined by the 2,4-dinitrophenylhydrazine procedure.34 Cytosolic protein thiols were determined following Sedlak and Lindsay.35 Glutamine synthetase activity was determined by the method of Stadtman etal.36
Protein concentration was determined by the Bradford method using Biorad reagent with bovine serum albumin as standard.37
Statistical Analyses
Data are expressed as means ± SEM. Statistical differences between groups were calculated by the Student unpaired t test or,for multigroup comparisons, by ANOVA, followed by Bonferroni’spost-hoc test. The relationship between normaly distributed variables was assessed using Pearson’s correlationcoefficient.
RESULTS
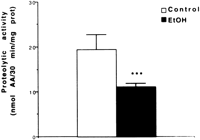
Liver protein concentrations (206.9 ± 17.3 vs. 223.8 ± 18.6 mg/g wet liver tissue; n = 6) were not significantly different in pair-fed controls and ethanol-infused rats. Cytosolic alkaline protease activity was determined in the liver of rats fed intragastrically, using intracellular proteins as substrates. This assay should provide data that more closely reflects the in vivo situation. As shown in Fig. 1, a significant decrease (![]() 43%; P < .01) in the alkaline protease activity was evident in the ethanol-fed rats as compared with pair-fed controls. Our results suggest that either proteins altered by oxidative stress in vivo are not good substrates for alkaline proteases proteolysis, and/or that these proteases are deficient or defective in ethanol-fed rats.
43%; P < .01) in the alkaline protease activity was evident in the ethanol-fed rats as compared with pair-fed controls. Our results suggest that either proteins altered by oxidative stress in vivo are not good substrates for alkaline proteases proteolysis, and/or that these proteases are deficient or defective in ethanol-fed rats.
| Fig. 1. Alkaline protease activity in hepatic cytosolic fractions of rats fed intragastrically with ethanol (EtOH) or an isocaloric amount of dextrose (control). Values are means ± SEM for four rats in each group; each determination was performed at least in duplicate. Statistically significant differences between groups were determined by the Student t test. ***P < .01 vs. controls. |
To further investigate this latter possibility, we determined two of the characteristic peptidase activities of the SDS-activatable 20S proteasome, the major components of the alkaline proteases, using specific fluorogenic peptides as substrates.
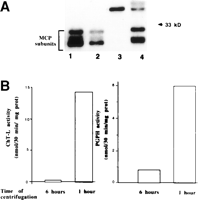
First, we checked that the ChT-L and PGPH activities, when measured in cytosolic fractions, were specifically related to the MCP (Fig. 2). We performed a Western blot analysis of the proteasome and determined the ChT-L and PGPH activities in supernatant fractions obtained by centrifugation at 100,000g for 1 or 6 hours. Western blot analysis (Fig. 2A) revealed that MCP subunits (21-32 kd) were detectable only in the 1-hour-centrifuged cytosolic fraction. The measurement of peptidase activities demonstrated that the fluorogenic peptides were degraded almost exclusively in the 1-hour-centrifuged cytosolic fraction (Fig. 2B). These results indicated that peptidase activities measured in the cytosolic fractions were almost exclusively related to the proteasomes.
| Fig. 2. SDS-polyacrylamide gel electrophoresis of proteasome and peptidase activities. (A) Western blotting was performed using rabbit anti-rat proteasome antiserum as described in Materials and Methods. Lanes 1-2: purified rat liver proteasome (0.5 and 0.25 µg, respectively). Lane 3: 75 µg of proteins from supernatant obtained after the centrifugation of liver homogenate from control rats at 100,000g for 6 hours. Lane 4: 75 µg of proteins from supernatant obtained after the centrifugation of liver homogenate from control rats at 100,000g for 1 hour. A molecular-weight marker (33 kd) is indicated on the right side. (B) Peptidase activities were determined using fluorogenic peptides as described in Materials and Methods, in the same supernatants used in Western blotting, lanes 3-4. |
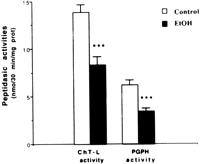
Next, we determined proteasome peptidase activities in rats fed intragastrically. Our data showed that ChT-L and PGPH activities were both significantly decreased in the liver of ethanol-fed rats (Fig. 3). It is interesting to note that the decrease inChT-L and PGPH activities was still significant when activitieswere measured either in postnuclear or postlysosomal fractions(results not shown).
|
|
Fig. 3. Peptidase activities in hepatic cytosolic fractions of rats fed intragastrically with ethanol (EtOH) or an isocaloric amount of dextrose (control). Values are means ± SEM for 10 rats in each group; each determination was performed at least in duplicate. Statistical differences between groups were determined by the Student t test. ***P < .001 vs. respective control. |
Samples assayed in the absence of SDS showed lower peptididyl activities, but the decrease in ethanol-treated rats was maintained. In fact, ChT-L activity was still significantly diminished (control:1.45 ± 0.42 nmol/30 min/mg protein; ethanol: 0.76 ± 0.17 nmol/30 min/mg protein; P < .05), indicating that ethanol treatment alsodecreased the basal activities of the proteasome.
No activation of peptidase activities was found when cytosolic fractions were dialyzed against Tris–ethylenediaminetetraacetic acid buffer, suggesting that the ethanol-related decline in peptidase activities is not related to a reversible inhibitor of molecular mass less than 12 kd (results not shown).
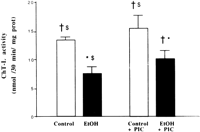
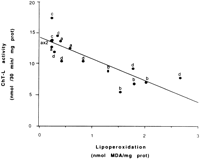
Among the mechanisms implicated in the decline in proteasomal peptidase activities, reactive oxygen species and lipid peroxidationby-products appear to play an important role. We measured theChT-L activity in animals treated with ethanol plus PIC, an inhibitor of CYP2E1. Treatment of control rats with PIC did not significantly modify the ChT-L activity. However, in rats receiving ethanol plus PIC, the ethanol-related inhibition was significantly reduced (Fig. 4). Most of these rats had higher peptidase activities and lower lipoperoxidation compared with the rats receiving ethanol alone. When peptidase activity was plotted against lipoperoxidation, an inverse correlation (r = ![]() .840; P < .01; n = 17) was observed between the individual values of ChT-L activity and those of lipoperoxidationlevel (Fig. 5). Moreover, CYP2E1 protein was inversely correlated with ChT-L activity (r = .901; P < .01; n = 17).
.840; P < .01; n = 17) was observed between the individual values of ChT-L activity and those of lipoperoxidationlevel (Fig. 5). Moreover, CYP2E1 protein was inversely correlated with ChT-L activity (r = .901; P < .01; n = 17).
| Fig. 4. ChT-L activity in hepatic cytosolic fractions of rats fed intragastrically with dextrose (control), ethanol (EtOH), dextrose + PIC (control + PIC), or ethanol + PIC (EtOH + PIC). Values are means ± SEM for three to five rats in each group; each determination was performed at least in duplicate. Statistically significant differences between groups as determined by ANOVA and the Bonferroni post-hoc test are indicated as: *P < .002 vs. control; |
| Fig. 5. Correlation between lipid peroxidation and ChT-L activity in liver of rats fed intragastrically with dextrose (a), ethanol (b), dextrose + PIC (c), or ethanol + PIC (d). r = |
To verify the role of free radical processes in the decrease of proteolysis, we determined peptidase activities and cellular free radical targets in livers obtained from rats receiving chronic ethanol as sole drinking fluid. The results showed that ChT-L and PGPH activities were not significantly different in 4-week ethanol-treated and control animals. The same negative results held true for the lipid peroxidation level, the cytosolic protein thiols, the protein carbonyl groups, and glutamine synthetase activity (table 1).
| Effects of Oral Ethanol Administration on Cellular Free Radical Targets and Peptidase Activities in Rat Liver |
DISCUSSION
Our present data indicate that alkaline protease activity declines in the liver of rats fed intragastrically with ethanol for 1 month. Alkaline proteases have been specifically implicated in the degradation of oxidatively modified forms of some proteins.38 In the livers of the same rats used in the present study, we showed that ethanol elicits an increased protein oxidation as demonstrated by the decrease in glutamine synthetase activity and in protein thiol groups, in addition to an increase in protein carbonyl groups.22 Surprisingly, such an increased supply of endogenous potential substrates for the cytosolic degradative system did not appear to enhance the activity of alkaline protease. The decrease in alkaline protease could be partly caused by the decrease in the 26S proteasome-related ubiquinated protein pathway, because ubiquitin conjugates and ubiquitin mRNA were shown to be reduced in the intragastric model.39 However, several studies have reported an accumulation of oxidized proteins and a decline in protease activity toward exogenous substrates in rat liver40,41 during aging. Because the 20S proteasome is primarily involved in the degradation of oxidized proteins, we studied this pathway, i.e., we used exogenous synthetic substrates to determine peptidase activities of the 20S proteasome, a major component of alkaline protease.
Whereas no change in two of the peptidase activities of the proteasome, ChT-L and PGPH activities, was observed in the oral ad libitum ethanol administration model, a decrease was already apparent in these activities in rats receiving ethanol intragastrically for 1 month. Interestingly, Donohue et al.42 reported that chymotrypticactivity was reduced after 2 months of intragastric ethanol infusion. Taken together with our results, these observations indicate that, in the intragastric ethanol infusion model, the impairment in cellular adaptation to oxidative stress as manifested by an increase in oxidized proteins and a dearth in antioxidants,22 implies the decrease in proteasomal peptidase activities. In contrast, the oral ethanol administration model was not associated with changes either in cellular targets of free radicals or in changes in proteasomal peptidase activities. It appears thus that the decrease in proteolysis is associated with the severity of ethanol-related oxidativestress.
Because the proteasome peptidase activities are still diminished in the presence of good synthetic substrates, it may be that oxidatively damaged proteins, which appear resistant to degradation, inhibit the intracellular proteases. In fact, oxidated ethanol-related protein modifications such as carbonyl groups could be the cause of the inhibition, because several small peptides that are proteasomeinhibitors contain aldehydic functions.43,44 Alternatively, reactive oxygen species and lipid peroxidation by-products could also be implicated in the inhibition of the proteasome. In fact, induction of CYP2E1 by ethanol has been postulated to cause hepatotoxicity by formation of radical intermediates and activated oxygen species.26,45Because the inhibition of CYP2E1 induction by coadministrationof PIC with ethanol resulted in an increase in ChT-L activitycompared with ethanol alone, it appears that reactive oxygen speciescould contribute to the inhibition of ChT-L activity. In fact,it has been reported that some peptidase activities of the proteasome purified from rat liver could be inactivated by the mixed-function oxidase system.32,46 Moreover, the inhibition of CYP2E1 expression by PIC strongly decreased lipid peroxidation,26 suggesting that lipid peroxidation products may also act to inhibit the proteasome. In fact, our data show that neither the lipoperoxidation level nor the ChT-L and PGPH activities are changed in the oral ethanol administration model, whereas an overt increased lipid peroxidation22 associated with a decrease in peptidase activities is observed in the intragastric infusion model. Moreover, there is a good correlation between the extent of lipid peroxidation and the changes in peptidase activity in the intragastric infusion model. Although such results do not demonstrate a causal relationship, these data are consistent with the proposal that severe oxidative stress could decrease proteolysis10 and that this highlights a possible role for lipoperoxidation by-products.
Lipid peroxidation may lead to the production of toxic and reactive aldehyde metabolites. Among these, MDA and 4-hydroxy-2-nonenal (HNE) are the most important. Using the intragastric infusion model, Tsukamoto et al.47 showed that products of lipid peroxidation are increased in the liver of ethanol-fed rats. These highly cytotoxic metabolites could form covalent links with various molecules, such as proteins. Using HNE adducts formed with glyceraldehyde phosphate dehydrogenase, Friguet et al.48,49 showed in in vitro studies that these cross-linked proteins are poor substrates for the proteasome, partly because the multicatalytic enzyme may be unable to bind large molecular material; alternatively, they may act as a potent inhibitor of the proteasome. Such cross-linked proteins, detected as immunoreactive adducts formed with MDA or HNE, are increased during ethanol treatment in micropigs,50rats,26,50 or patients.50-52 Moreover, the binding of acetaldehyde, the first metabolite of ethanol, to cytosolic proteins could also lead to the formation of covalent protein adducts.50 One can thus suggest that cross-linked proteins may also account in vivo, at least in part, for the decrease in proteasome activity occurring in ethanol-fed rats.
An inefficient removal related to the decreased activity of the 20S proteasome, the catalytic core of the 26S proteasome, may result in the accumulation of damaged proteins and in alterations in turn-over of proteins, and these can be associated with cytotoxicity. For example, the ethanol-related decrease in proteasome activities may contribute to the decrease in CYP2E1 turn-over and the subsequenthigh level of CYP2E1 achieved in the intragastric infusion model,21 in view of the fact that CYP2E1 is a substrate for ubiquitin and/or proteasome-mediated proteolysis.53-56 Also, because the ubiquitin-proteasome system located in the cytoplasm has emerged as the major endoplasmic reticulum degradation machinery,57 the decrease in the rateof microsomal protein degradation described by Tobon and Mezey13 could, at least partly, be related to changes in the proteasome pathway, and thus may contribute to the ethanol-induced accumulation of hepaticproteins.
In conclusion, we show that the chronic intragastric ethanol infusion decreases alkaline protease activity, probably through the inhibition of ChT-L and PGPH activities of the proteasome. The mechanism of inhibition could involve aldehyde-derivative peptides, reactive oxygen species, or cross-linked proteins, all of them being generated following chronic extensive oxidative stress. Whatever the exact mechanism, it appears that a failure in the proteasome pathway to clear the hepatocytes of damaged proteins results from a chronic cellular oxidative stress. The inhibition of the proteasome pathway appears to be involved in the oxidatively challenged alcoholic livers and may play a pathogenic role in experimental alcoholic liver disease.
Acknowledgment
The authors thank Dr. B. Friguet for the kind gift of MCP and antisera against MCP. They are grateful to Pr. Ingelmann-Sundberg for providing data on CYP2E1.
Abbreviations:
MCP, multicatalytic protease; ChT-L, chymotrypsine-like; PGPH, peptidylglutamyl peptide hydrolase; SDS, sodium dodecyl sulfate; CYP2E1, cytochrome P4502E1; PIC, phenethyl isothiocyanate; MDA, malondialdehyde; HNE, 4-hydroxy-2-nonenal.
Footnotes
Received April 17, 1998; accepted July 29, 1998.
Supported by grants from the Université René Descartes, Paris, France, from IREB (98/20), and from NIH-NIAAA 8116, Bethesda,MD.
Address reprint requests to: Prof. Hélène Rouach, Lab. of Biomedical Research on Alcoholism, 45 rue des Saints-Pères, 75270 Paris Cedex 06, France. Fax: 33-1-42-86-04-02.
REFERENCES