HEPATOLOGY, February 1998, p. 621-627, Vol. 27, No. 2
Original Articles
Full-Length Complementary DNA of Hepatitis C Virus Genome From an Infectious Blood Sample
Hideki Aizaki1, Yoichiro Aoki1, Takashi Harada1, Koji Ishii1, Tetsuro Suzuki1, Seishi Nagamori2, Gotaro Toda2, Yoshiharu Matsuura1, and Tatsuo Miyamura1
From the 1 Laboratory of Hepatitis Viruses, Department of Virology II, National Institute of Infectious Diseases, Toyama, Shinjuku-ku, Tokyo, and 2 Internal Medicine(I), Jikei University School of Medicine, Nishishinbashi, Minato-ku, Tokyo, Japan
ABSTRACT
We constructed a full-length complementary DNA (cDNA) clone of Hepatitis C virus (HCV) from a blood sample of an HCV carrier. The blood from the carrier was eventually transfused to a patient who later developed typical posttransfusion Hepatitis C. It was also shown to be infectious to chimpanzees. We obtained 12 overlapping cDNA fragments altogether, covering the entire HCV genome. By subcloning and sequencing, clones considered to constitute the major population were selected. We could also detect 98 base pairs of extra sequences at the 3′ end of the genome. After confirming the overlapping sequences, we combined the fragments to make a full-length cDNA. The HCV population in the donor was heterogeneous, as determined by their nucleotide sequences of the hypervariable region in envelope protein, but a few virus clones were selected in the recipient after transmission. The similar convergence of the virus population was previously observed when the same blood sample was injected into a chimpanzee. Interestingly, virus clones isolated during the acute phase in the recipient and the chimpanzee had sequences in the hypervariable region identical to that of the full-length cDNA clone. The full-length cDNA clone of HCV constructed in this study may originate from infectious virus clones.(HEPATOLOGY 1998;27:621-627.)
INTRODUCTION
Comparative study of the amino acid sequence of Hepatitis C virus (HCV) with those of flaviviruses and pestiviruses and gene expression experiments in bacteria, yeast, and animal cells have revealed that the proteins of HCV are processed by a host- derived signalase and cleaved by virus-coded proteases.1-5 In vitro culture systems that support partial replication of this virus have been developed from human T- and B-cell lines, 6,7 human fetal hepatocytes,8chimpanzee hepatocytes,9 and human primary hepatocytes.10 However, fully efficient, long-term viral replication has not yet been established. To study the biology of HCV and its pathogenesis, it is essential to establish an efficient in vitro cell culture system or an infectious complementary DNA (cDNA) clone to support the complete replication of HCV.
As a first step to construct an infectious cDNA clone of HCV, it is important to select an adequate sample containing infectious HCV. To date, a considerable number of “complete” clones of the HCV genome have been reported. However, it is not certain that those clones have truly originated from infectious HCV, because the materials used usually were pooled plasma samples. Furthermore, the plasma of HCV carriers or patients is generally composed of quasispecies of HCV population.11 To construct an infectious cDNA clone of RNA viruses, it is crucial to retain both 5′ and 3′ ends of the sequence, which are highly conserved and are considered to play essential roles for RNA synthesis, transcription, and translation.12 Many have reported that the 3′ untranslated region (UTR) of HCV consisted of poly(U)13-16 or poly(A)17 homopolymer tracts. A novel 98-nucleotide (nt) sequence downstream from the poly(U) stretch of the HCV genome was recently reported. 18,19
To construct a complete cDNA clone of the HCV genome, we selected a blood donor sample which was shown to be infectious to both humans and chimpanzees.20-23 From that sample we have cloned several fragmental cDNAs and constructed a full-length cDNA containing the 98-base pair (bp)-3’X sequence. We also compared the sequence in the hypervariable region (HVR) of HCV in the donor from whom the cDNA was derived with those derived from the recipient and the experimentally infected chimpanzee.
MATERIALS AND METHODS
Material. We selected a donor blood sample that had been shown to be infectious to both human and chimpanzees.20-23 This plasma was obtained from a healthy blood donor who was implicated in the transmission of HCV. The donor blood was later found to be positive for anti-HCV antibody. The genomic RNA measured by reverse transcription-polymerase chain reaction (RT-PCR) of the plasma was 105 genome/mL. One milliliter of 105 dilution of the plasma made a chimpanzee develop hepatitis, as demonstrated by elevated serum alanine transaminase (ALT) levels, histological changes on liver biopsies, detection of anti-C100 HCV antibody, and the presence of HCV RNA. It has been shown that the 50% chimpanzee infectious dose of the plasma was 105.5CID50/mL. 22,23
Clinical Characteristics of the Recipient Patient. One patient developed typical posttransfusion Hepatitis C after he underwent surgery for empyema. The patient had elevated levels of serum transaminase, antibodies against C-100, P-22 antigens, and plasma HCV RNA. The diagnosis of acute hepatitis was confirmed by liver biopsy. The serum of the patient was negative for Hepatitis B surface antigen. The patient was treated with 0.75 million U/d of interferon alfa (IFN-![]() ) intramuscularly seven times a week for 2 weeks and three times a week for 6 months. Plasma samples were obtained serially and stored at
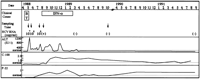
) intramuscularly seven times a week for 2 weeks and three times a week for 6 months. Plasma samples were obtained serially and stored at ![]() 80°C. The clinical profile of the patient is shown in Fig. 1. The time courses of ALT elevation, anti-HCV antibody levels (anti-C-100 and anti-P22), HCV RNA titer, and sampling times are also illustrated. HCV RNA titer was measured by competitive PCR.24 When the titers were less than 103 genome Eq/mL, the presence of HCV RNA was examined by RT-nested PCR using the 5′ UTR.25
80°C. The clinical profile of the patient is shown in Fig. 1. The time courses of ALT elevation, anti-HCV antibody levels (anti-C-100 and anti-P22), HCV RNA titer, and sampling times are also illustrated. HCV RNA titer was measured by competitive PCR.24 When the titers were less than 103 genome Eq/mL, the presence of HCV RNA was examined by RT-nested PCR using the 5′ UTR.25
|
|
Fig. 1. Clinical characteristics of the recipient. The profiles of ALT values (IU/L) and anti-HCV antibody (C-100 and p-22) titers in the recipient from March 1988 to September 1991 are shown. Open squares indicate the periods of blood transfusion (BT) and IFN treatment (IFN- |
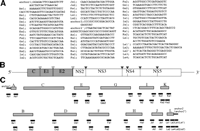
RNA Preparation, cDNA Synthesis, and PCR. RNA was extracted from 80 mL of the plasma using the guanidium/cesium chloride method described previously.26 The cDNA was synthesized with 200 units of Superscript II (BRL, Gaithersburg, MD) by using antisense primers. The cDNA was amplified by 30 cycles of nested PCR (90 seconds at 94°C, 90 seconds at 55°C, 2 minutes at 72°C) by using both sense and antisense primers. The primer sequences are illustrated in Fig. 2 A.
|
|
Fig. 2. Construction of a full-length cDNA clone of HCV. (A) Sequences of primers and anchors for RT-nested PCR. The synthetic oligonucleotide primers are according to the published nucleotide sequences of type Ib HCV clones and a novel 98-nt sequence in the 3′ terminus. The anchors (anchor 1, anchor 2′) and the anchors’ primers (As1, La1′, La2′) are obtained from the 5′-Ampli FINDER RACE kit and Marathon cDNA amplification kit. An oligo RNA (anchor 2′) and primers (Ls1, Ls2, Ls3, Ls4, La1, La2) were according to Tanaka et al.63 (B) Gene organization of the HCV. Shaded and open boxes indicate structural and nonstructural proteins, respectively. UTRs are indicated by bars. Black and white triangles indicate cleavage sites by cellular signalase and viral-coded proteases, respectively. (C) Strategy for making a full-length cDNA clone of HCV. The locations of primers and anchors are shown. The 5′ and 3′ terminal sequences were determined by use of a 5′-Ampli FINDER RACE kit and an RNA linker ligation, respectively. |
To clone the cDNA in the 5′ UTR, we adopted the 5′ rapid amplification of cDNA ends method27-29 using the 5′-Ampli FINDER RACE Kit (Clontech, Palo Alto, CA). The cDNA was synthesized with the antisense primer Aa1 (Fig2A), and the RNA template was hydrolyzed with NaOH. After neutralization, excess primer was removed and the cDNA was concentrated by ethanol precipitation. A single-stranded anchor 1 oligonucleotide was ligated to the 3′ end of the cDNA, and the anchor-ligated cDNA was used as a template for 40 cycles of PCR amplification (1 minute at 94°C, 1 minute at 55°C, 2 minutes at 72°C) by using a pair of primers, As1 and Aa1 (Fig.2 A).
To clone the 3′ end of the genome from the serum by oligo RNA ligation, the phosphorylated oligo RNA (anchor 2 in Fig. 2 A) was synthesized (Takara Shuzo, Kyoto, Japan). The RNA prepared from 80 mL of the serum and the 5′-phosphorylated oligo RNA were ligated by T4 RNA ligase (Takara) at 10°C for 15 hours according to the manufacturer’s protocol. RT was performed at 50°C for 1 hour with primer La1 (Fig. 2 A). The RT product was amplified by nested PCR with the following primers: first PCR; Ls1 and La2; 2nd PCR; Ls2 and La2.
For isolation of the 5′ end of the antigenome, we used modified 3′ RACE method (Marathon cDNA Amplification Kit [Clontech]). The antigenomic strand of HCV RNA was synthesized by the primer Ls1 with the reverse transcriptase of Moloney murine leukemia virus (Fig. 2 A). The cDNA was treated with a conventional cocktail of Escherichia coli DNA polymerase I, RNase H, and E.coli-DNA ligase according to the method for generating cDNA libraries.30 After creating the blunt ends with T4 DNA polymerase, the double-stranded cDNA was ligated to the anchor 2′ sequence. A set of primers (first PCR; Ls1 and La1′, second; Ls2 and La2′; Fig. 2A) was applied for 40 cycles of nested PCR (45 seconds at 94°C, 45 seconds at 55°C, 2 minutes at 72°C).
Cloning and Sequencing. The amplified cDNA fragment was purified from an agarose gel and treated with T4 polynucleotide kinase for phosphorylation. The fragment was cloned by blunt-end ligation into the SmaI site of pUC119 (Fig. 2C). The nucleotide sequence was determined using the Taq dideoxy terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, CA) after various deletions with the kilosequence deletion kit (Takara Shuzo). We isolated several clones in each region and determined their nucleotide sequences in both strands. The clone that constituted the major population was used to make the full-length cDNA clone. The overlapping regions of the clones were compared with those of adjacent clones.
Construction of a Full-Length cDNA Clone of HCV. The clones pUCK and pUCL (Fig. 2C) were jointed to make pUCKL by use of the PCR method with the primer Ks2 and La2 after heat shock (70°C for 15 minutes) of the mixture of pUCK and pUCL. These cDNA clones were then combined to a full-length clone of HCV NIHJ1 by shared restriction enzyme site, namely, pUCA HindIII (0) – NcoI (83), pUCB NcoI (83) – ClaI (708), pUCC (modified from pSR327)31 ClaI (708) – SacI (2322), pUCD SacI (2322) -SacII (3310), pUCE SacII (3310) – NdeI (4069), pUCF NdeI (4069) – BstEII (5243), pUCG BstEII (5243) -EcoRI (6699), pUCH EcoRI (6699) – PflMI (7528), pUCI PflMI (7528) – PstI (8497), pUCJ PstI (8497) -BspMI (9355), pUCKL BspMI (9355) – HindIII(98′) and replaced into HindIII site of pBR322 vector (Fig. 2C). The nucleotides in the 3′ extra sequence were numbered 1′ to 98′.
Determination of the 3′ Extra Sequence in the Plasma of Donor and Recipient. To analyze the existence of the 3′ extra sequence of HCV, we determined the sequences of positive- and negative-stranded RNA of HCV in the plasma of the donor and recipient during clinical phases (Fig. 1). In addition, an RT reaction was carried out in the absence of the antisense primer with Superscript II after chemical modification of RNA to analyze the specificity of negative-stranded HCV RNA.32 RT reaction was conducted by use of the primer La3 (for positive-stranded RNA) and primer Ls1 (for negative-stranded RNA) (Fig.2A). The reaction mixture was heated at 100°C for 30 minutes to inactivate the reverse transcriptase, and two sets of primers (first PCR; Ls1 and La3, second; Ls2 and La4) were used for 40 cycles of nested PCR (45 seconds at 94°C, 45 seconds at 55°C, 2 minutes at 72°C). PCR products were subcloned and sequenced as described above.
Analyses of HVR of Clones Recovered From the Donor and Recipient. The sequence variation of the HVR during the clinical course was examined. RNA extracted from each time point was amplified by RT-PCR with specific primer pairs for the HVR.33 PCR products were subcloned, and nucleotide sequences of 10 clones were determined. The sequences of these clones were compared with that of the NIHJ1 and those obtained from an experimentally infected chimpanzee.
Phylogenetic Analysis. To determine the heterogeneity of the virus isolated from the donor, recipient, and chimpanzee, phylogenetic trees of HVR of the 15 HCV isolates were constructed by the neighbor-joining method34 using the number of synonymous substitution35 with the Clustal W program.36
RESULTS
Identification of an Implicated Donor. In April 1988, a healthy person donated blood. Including this donor’s blood, a total of 6 units of blood was transfused to a patient who underwent surgery. After 4 weeks, the recipient developed posttransfusion Hepatitis C, the clinical course of which is illustrated in Fig. 1. A 6-month treatment of IFN-![]() (0.75 MU/D, 3 times a week) produced dramatic results. HCV RNA could not be detected after the ALT level became normalized and has not been detected since. The implicated donor was retrospectively identified by the presence of anti-HCV antibody (anti-C100). All other donor blood was negative for the HCV antibody. Approximately 1 year after the transfusion, blood was obtained from the donor. HCV cDNA fragments were obtained from the blood by RT-PCR. 20,21 Blood collected 6 months later was injected into chimpanzees and was infectious. The infectivity was 105.5chimpanzee infectious doses/mL.23
(0.75 MU/D, 3 times a week) produced dramatic results. HCV RNA could not be detected after the ALT level became normalized and has not been detected since. The implicated donor was retrospectively identified by the presence of anti-HCV antibody (anti-C100). All other donor blood was negative for the HCV antibody. Approximately 1 year after the transfusion, blood was obtained from the donor. HCV cDNA fragments were obtained from the blood by RT-PCR. 20,21 Blood collected 6 months later was injected into chimpanzees and was infectious. The infectivity was 105.5chimpanzee infectious doses/mL.23
Construction of a Full-Length HCV cDNA From the Infectious Blood Sample.Twelve overlapping cDNAs covering the entire HCV genome were amplified by the PCR from the donor blood sample (Fig. 2B). We then subcloned 10 of each clone from these 12 regions and determined their respective sequences. Their overlapping regions had sequences almost identical to those of their adjacent clones. A plasmid containing a full-length cDNA clone of HCV, designated pNIHJ1, was constructed by ligation of each of 12 clones representing the major population into the HindIII site of pBR322. The pNIHJ1 consists of 341 nt of 5′ UTR, 9,033 nt of single open reading frame (ORF), 38 nt of 3′ UTR, 38 nt of poly(U) stretch, and 98 nt of the 3′ extra sequence (3’X). The base composition of this sequence was 20.3% adenine, 21.5% thymine, 30.0% cytosine, and 28.2% guanine.
Determination of 3’X Sequence in the Plasma of Donor and Recipient. We compared the 3’X sequence of pNIHJ1 clone with those reported previously.18,19,37 Sequence homology of the 3’X of pNIHJ1 against other type 1b clones and different types were 97% and 96%, respectively. These results confirmed that the 3’X sequences were highly conserved among different HCV isolates.
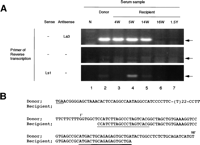
In the recipient plasma collected 4 to 14 weeks posttransfusion, the 3’X sequence was consistently positive (Fig. 3A, lanes 3-5). However, immediately after the IFN therapy (16 week posttransfusion), it turned negative (Fig. 3A, lane 6). The 3’X sequence at 4 weeks posttransfusion was identical to the pNIHJ1 (Fig. 3B). We also analyzed the 5′ end sequences of the negative strand of the HCV genome. Although the band was not detectable at 4 weeks posttransfusion, a clear band was detected at 5 weeks, was still detectable at 14 weeks, and disappeared after IFN therapy. We detected no band in the same RT condition without primers.
|
|
Fig. 3. Detecting and sequencing of the 3′ terminal sequence of HCV from donor and recipient plasma. (A) Detection of the terminal extra sequence in positive and negative strands of HCV RNA and comparison with the presence of 5′ UTR sequence. The presence of the 3′ extra sequence was examined by RT-nested PCR and electrophoresed on a 3% agarose gel as described in Materials and Methods. The primers used in RT are indicated at the left. Arrows indicate the expected size of RT-PCR products. Lane 1, normal serum (N); lane 2, plasma sample of the donor; lane 3, recipient sample 4 weeks posttransfusion (4W);lane 4, 5 weeks posttransfusion (5W); lane 5, 14 weeks posttransfusion (14W); lane 6, 16 weeks posttransfusion (16W); and lane 7, 1.5 years posttransfusion (1.5Y). (B) Homology analysis of the 3′ terminal sequence of HCV derived from donor and recipient. The upper sequences represent the 3′ terminal sequence of pNIHJ1. The lower sequences indicate the terminal sequences obtained from the recipient 4 weeks posttransfusion. The nucleotides in the 3′ extra sequences are numbered 1′ to 98′ above the sequences. The termination codon of the ORF (TGA) and sequences of the primers used in PCR are underlined. |
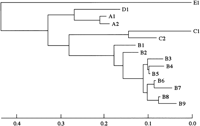
Analyses of HVR of Clones Recovered From Donor and Recipient. To examine the HCV population after transmission, we compared the sequences of HVR of 15 HCV clones obtained from the donor, the experimentally infected chimpanzee, and the recipient at several clinical stages (table 1). These virus clones were classified into 5 major groups (A-E) by phylogenetic tree analysis (Fig. 4). Although the A1 clone was the major clone (18/30) in the donor population, the B1 clone became dominant in the inoculated chimpanzee (19/20).38 On the other hand, virus clones with different sequences were recovered from the recipient in the early stage of infection (4 weeks posttransplantation). The population converged gradually to one clone, the B8 clone. After IFN therapy (16 weeks posttransplantation), only the E1 clone, which was not detected in the original donor blood, was isolated (10/10).
| View This table | table 1. Sequences of HVR and Number of Clones Recovered |
|
|
Fig. 4. Phylogenetic analysis of HVRs of the virus clones recovered from the donor, the recipient, and the chimpanzee. Phylogenetic trees of 15 HCV clones recovered from the donor, the recipient, and the chimpanzee were constructed based on the sequences of HVR by the neighbor-joining method. Horizontal lines indicate the nucleotide sequence distance between the sequences. The virus clones were classified into 5 major groups (A-E). |
The pNIHJ1 clone has an amino acid sequence in HVR identical to that of the A1 clone, which was the major clone in the donor sample and also was recovered from the recipient and chimpanzee after inoculation. This might indicate that the pNIHJ1 clone is derived from HCV clones infectious to both chimpanzees and humans.
DISCUSSION
Immediately after the prototype viruses were cloned and identified, more than 20 complete sequences of HCV clones were reported. 13-16,39-47 In most cases, the starting materials were plasma pools of donated blood. The cDNA fragments were obtained by RT-PCR using many sets of primers derived from the prototype HCV sequence. Therefore, it is not certain that the obtained sequences originated from infectious HCV particles. No sufficient system to evaluate the replication of HCV in vitro or to clone an infectious particle from the quasispecies population of HCV is available yet. Therefore, it is impossible to assess the authenticity of the cDNA clones of HCV at the moment. However, the complete cDNA clone of HCV constructed in this study is believed to originate from infectious HCV clones for the following reasons: 1) the blood sample from the donor was infectious to both chimpanzees and humans 20,22 ; 2) from the sample, we isolated 12 overlapping clones covering the entire region of HCV that were major clones in each region, and their overlapping sequences were almost identical to the adjacent clones; and 3) the virus clones with sequences in HVR identical to those of the complete cDNA clone were detected in both recipient and chimpanzee after infection. Recently, Kolykhalov et al. showed that in vitrosynthesized full-length HCV RNA derived from the consensus cDNA clone was infectious to a chimpanzee after direct injection into the liver.48 In our study, the consensus cDNA clone was constructed by choosing the major clone in each region.
One of the problems in constructing an infectious cDNA clone of viruses is the instability of cDNA clones in bacteria. To establish full-length infectious cDNA clones of yellow fever virus49 and Japanese encephalitis virus50 successfully, it was necessary to elude mutations during propagation of the plasmid in bacteria. In this study, to minimize this possibility, we used a recombination negative E.coli strain (DH10B) and low copy number plasmid (pBR322) in the construction of the full-length cDNA clone. To confirm the accuracy of an ORF of our clone, we expressed the entire ORF in mammalian and insect cells, and properly processed proteins of core, E1, E2, nonstructural (NS) 2, NS3, NS4A, NS4B, NS5A, and NS5B proteins were detected (manuscript in preparation). Protease activity was detected in the recombinant proteins obtained by expression of NS3 region of this clone expressed in insect cells51 and bacteria.52 Furthermore, an RNA-dependent RNA polymerase activity was detected in the recombinant protein of NS5B region expressed in insect cells (manuscript in preparation).
Recently the 98-nt extra sequence downstream from the poly(U) stretch of the genomic strand of HCV RNA was reported. Tanaka et al.18 first detected the sequence by primer extension on the antigenomic strand RNA of HCV and by dC tailing of the cDNA. On the other hand, Kolykhalov et al.19 and Yamada et al.37identified the same sequence by use of an oligo RNA ligation technique. We also detected the 98-nt sequence at the 3′ end of the genomic RNA by an RNA linker ligation. All virus clones had the 98-nt sequences, except for some that had only 97-nt sequences. We verified the 3′ end sequence by using another method to clone the 3’X sequence from antigenomic RNA. This blunt ligation method was much more efficient than the original 3′ RACE method based on homopolymeric tailing or ligation of an adapter to single-stranded cDNA by T4 RNA ligase. By this method, we observed that the longest virus clones had the 98-nt sequence at the 5′ end of antigenomic RNA. The sequence was identical to that from the 3′ end of the genomic RNA (data not shown).
In this study, we demonstrated that an identical sequence of the 3′ terminus of the HCV genome was recovered from both donor and recipient blood. Furthermore, detection of the 3’X sequence was consistent with the viremic phase of the patient. Detection of the 3’X sequence was further confirmed by detection of 5′ extra sequence of the antigenomic strand. In fact, detection of the 5′ extra sequence of the negative strand is more sensitive, and this sequence was detected just after the ALT elevation. The terminal sequence in both strands disappeared soon after the IFN therapy. Indeed, the possibility still exists that the detection of the negative-stranded HCV RNA is an artifact caused by the high titer of positive-stranded HCV RNA in the sample.53 In this experiment, the titer of positive sense HCV RNA at 4, 5, and 14 weeks posttransfusion were 107,106, and 106, respectively. However, a strong band was detected only at 5 weeks posttransfusion in spite of the lower titer than that at 4 weeks posttransfusion. These data suggest that the presence of the 3’X is directly associated with the replication of HCV, and the 3’X is derived from infectious HCV virion. Recently, Yoo et al.54 reported that a full-length HCV RNA synthesized in vitro lacking the 3′ extra sequence was infectious to the human hepatoma cell line.54 In the report, a large amount of RNA transfection (2 mg/5 ×105 cells) yielded low levels of newly synthesized HCV RNA with no 3’X sequence. These results, unlike other reports, including our present study, may suggest that the 3′ extra sequence is dispensable for HCV replication. To address the significance of the 3’X, it is most important to compare the infectivity of HCV RNAs with or without the 3’X in cell culture systems that support the efficient HCV replication.
The HVR is thought to be the major targets of host immune response and to play an important role in escaping from neutralizing antibodies. 33,55,56 The full-length cDNA clone of HCV, pNIHJ1, had the same amino acid sequences as the virus clone A1, which was detected in both a human and a chimpanzee after infection. The infectivity of the clone A1 was also demonstrated in vitro in a cell culture system (HPB-Ma, AD-HPB, Daudi cell lines).38 Although many virus clones were detected in donor serum, the clones A1 and B1 were transmitted to both recipient and chimpanzee. These virus clones were free of antibody.38Similar mechanisms of selection seemed to occur in the human and the chimpanzee. However, this cannot explain why the B1 clone became dominant (19/20) in the early phase of infection in the chimpanzee, whereas a heterologous population persisted in the human recipient. As in cases of HIV infection, the quasispecies of infectious source of HCV converged to one clone, possibly by selection after transmission.57 In our study, the same source of HCV that contained infectious virus clones was transmitted to a human and a chimpanzee, and both developed hepatitis. The selection of viral population occurred in both hosts, and the resulting converged clone was different. Chimpanzees are the only experimental animal sensitive to HCV infection.58-60However, the immunological responses to HCV infection in humans and chimpanzees may be different. 26,61 In fact, the rate of transition to chronicity is much higher in humans than in chimpanzees.62 The data obtained in this study also suggest a difference in susceptibility to HCV between humans and chimpanzees.
The study of HCV has been hampered by the absence of an efficient in vitro replication system. Establishment of an in vitro replication system is an immediate necessity for HCV research. Our study indicates that the clone pNIHJ1 is derived from HCV clones infectious to both chimpanzee and human. In an independent study, we also confirmed that a complete ORF of the pNIHJ1 clone was capable of expressing all HCV proteins that were processed properly. The full-length cDNA clone of HCV constructed in this study is thus unequivocally useful in the study of HCV replication.
Footnotes
Acknowledgement: The authors thank Drs. M. Houghton and I. Saito for critical review; T. Katayama and H. Harada for clinical samples; S. Sugita for gene analysis; S. Morikawa and T. Katayama for cDNA cloning; and Y. K. Shimizu, H. Shimojo, H. Yoshikura, and A. Weiner for helpful discussion. They also thank Y. Hirama and A. Suzuki for technical assistance; and T. Mizoguchi for preparation of the manuscript.
Abbreviations: HCV, Hepatitis C virus; UTR, untranslated region; nt, nucleotide; HVR, hypervariable region; RT-PCR, reverse transcription-polymerase chain reaction; ALT, alanine transaminase; IFN, interferon; ORF, open reading frame; NS, nonstructural.
Supported in part by grants-in-aid from the Ministry of Health and Welfare; the Ministry of Education, Science and Culture; and the Basic Research Core System for the Agency of Science and Technology, Japan. H.A. is a research fellow of Japan Science and Technology Corporation.
Nucleotide sequence accession numbers: The sequences reported here have been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession number, D89815
Received July 21, 1997; accepted October 16, 1997.
Address reprint requests to: Tatsuo Miyamura, M.D., Department of Virology II, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162, Japan. Fax: 81-3-5285-1161.